 Faith & Science
Faith & Science
 Life Sciences
Life Sciences
Ayala and Falk Miss the Signs in the Genome
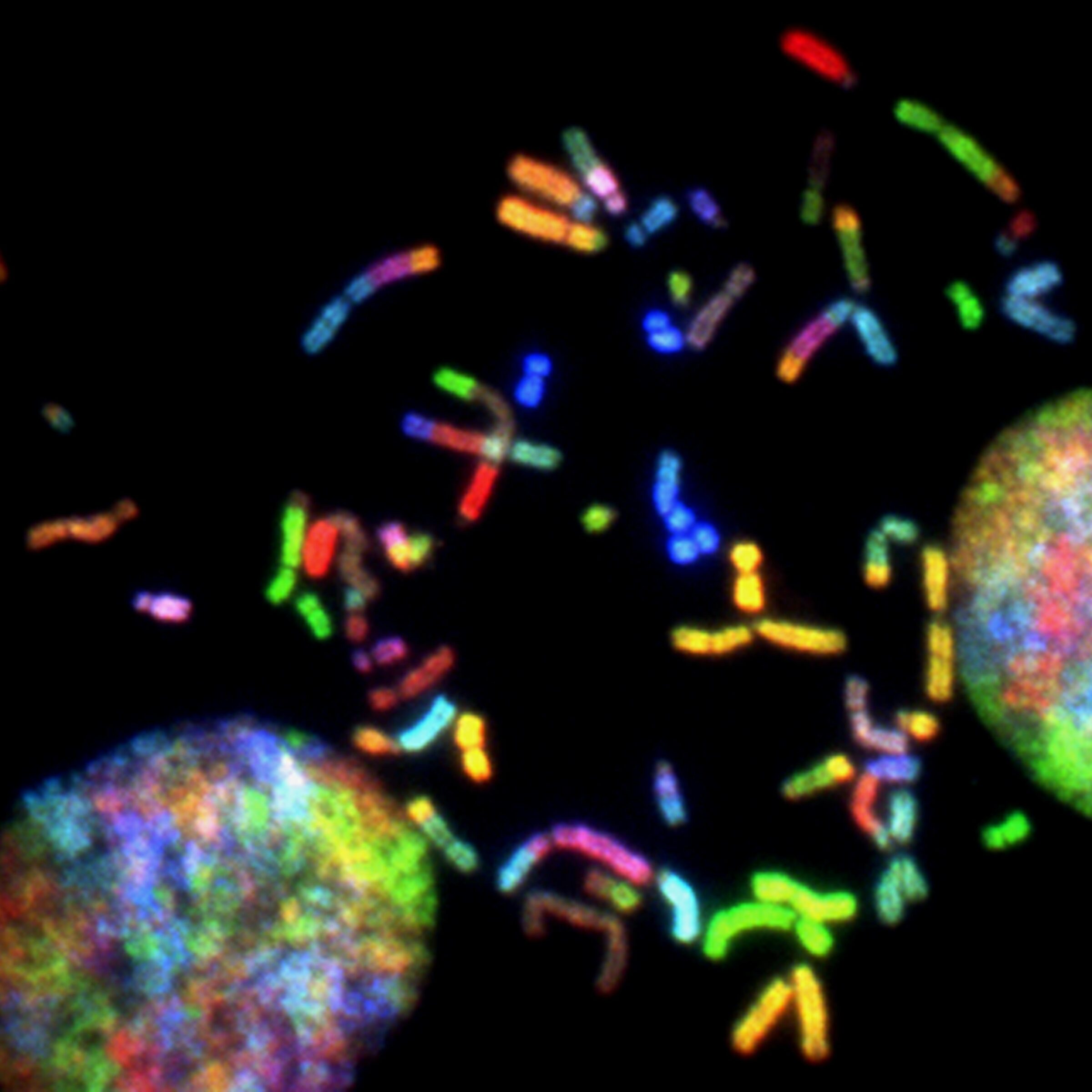
In his recent response to Stephen Meyer’s Signature in the Cell, Francisco Ayala claimed that repetitive portions of our DNA called “Alu” sequences are “nonsensical.” Ayala wrote: “Would a function ever be found for these one million nearly identical Alu sequences? It seems most unlikely.” In his response to Ayala, Meyer showed that Ayala is factually wrong about this. According to recent technical papers in genomics, Alu sequences perform multiple functions.
In a rejoinder to Meyer, Darrel Falk defended Ayala and claimed although “a number of functional regions have been discovered within Alu sequences,” there “is no question that many Alu sequences really have no function.”
In my last blog, I showed that the vast majority of the genome is transcribed, either into protein-coding genes or into regulatory RNAs. The technical literature — some of which I cited in that blog — reports that the genome is an RNA-coding machine. Clearly, most DNA really does have function.
In this and subsequent posts, I will provide other sorts of evidence that so-called “junk DNA” is not junk at all, but functional.
We have all seen a variant of the plot in a movie. A strange signal appears–in one film it is a recurrent wireless telegraph code that is transmitted from San Diego after a global nuclear holocaust (On the Beach); in another it is radio transmissions from deep space (Contact); in still another it is crop circles (Signs). As we all know, the first signal turns out to be due to a Coca-Cola bottle: Wind blowing on a window shade next to the bottle results in the latter being occasionally nudged, which sometimes leads to a telegraph key being tapped by the very same. But in the second movie, the signals received turn out to contain a complex set of encrypted data with an intricate mathematical pattern — they are the specifications for building a device that can travel through space-time wormholes, sent from a friendly alien civilization. So also are the crop circles in the third film messages from an extraterrestrial race, except that the designs portend an attack on humanity.
Now, the reason we are drawn in by such stories is obvious: The signals have serious implications for the characters. It could mean the survival of mankind after a thermonuclear war; it could mean that there are other sentient beings in the universe. That is why we would quickly lose interest in the plot if, say, in every scene where a scientist appeared before an important governmental group and said, “The outer space signal contains over sixty thousand, multidimensional pages of complex architectural plans,” she were countered with, “This is exactly the predicted outcome of billions of years of cosmic evolution — you see, random interstellar events lead to just this kind of complex specified information…we are not impressed.” We would want our money back.
My purpose for bringing up this subject is that I have a mysterious genomic signal for you to see — which I will show you tomorrow. We detected it some time ago and it has aroused the interest of some genomicists, but you will find no mention of it books such as Francis Collins’s The Language of God — which is peculiar. But I have another aim in mind, too, for broaching this possible chromosomal code: A key first indicator of functionality is a distinctly non-random pattern. The persistence of a distinct signal in different contexts often suggests functional constraints are operative–that is why genomicists look for them. And since I want to focus on the global functions of such Short Interspersed Nuclear Elements (SINEs) as human Alus and their mouse and rat counterparts, their far-from-random placement cannot be elided. In fact, I will argue that it is a critical part of the genome story that the folks at Biologos aren’t telling you.
To prepare for the mysterious genomic signal, though, I want to draw your attention to this figure:
What you are seeing are the relative densities of Long Interspersed Nuclear Element (LINE) L1s and SINEs along 110,000,000 DNA letters of rat chromosome 10.1 (From Fig. 9d of reference 1.) The x-axis represents the sequence of letters in DNA and the blue line indicates where SINEs occur — what Ayala calls “obnoxious sequences” that are supposedly due to “degenerative biological processes that are not the result of ID.” The red line indicates where LINE sequences occur.
By the way, Francis Collins is a principal author of the Nature paper where these results are published.
Both LINEs and SINEs are types of mobile DNA, namely, retrotransposons, and together they can comprise around half of the mammalian genome. As should be clear from the figure, LINEs tend to peak in abundance where SINEs taper off and vice versa (see the blue boxes). We have known about this pattern since the late 1980s, so it is no surprise to someone who has been following the subject. What should be surprising to anyone, however, is that the same machinery is responsible for the movement of both types of retrotransposon. A complete L1 element encodes the proteins necessary to “reverse transcribe” an RNA copy of itself back into DNA, and to insert the generated duplicate into some chromosomal site. SINEs, by way of contrast, rely on the L1-specified proteins for all their copying and pasting routines.
This compartmentalization of LINEs and SINEs along the mammalian chromosome can also be detected by using molecular probes for L1 or Alu(-like) sequences2:
For junkety-junk elements that can make up fifty percent of a mammal’s mostly junkety-junk genome, the rule seems to be: Location, location, location.
Interestingly, this higher-order pattern cannot be detected when small sections of DNA are examined. It only becomes evident when stretches that are millions of nucleotides long are studied.
This banding pattern has been known for decades–but for some reason it is rarely (if ever) discussed by “junk DNA” advocates. The bands on the chromosome arms fall into two general categories:
- R bands: DNA compartments that are enriched with the genetic letters G and C, have a high concentration of protein-coding genes, a preponderance of Alu or Alu-like repetitive elements (e.g., mouse B1s), and replicate early in the DNA synthesis phase of the cell cycle.
- G bands: DNA compartments that are enriched with the genetic letters A and T, have a low concentration of protein-coding genes, a high density of the L1 retrotransposon, and replicate late in the DNA synthesis phase of the cell cycle.
There are other strong functional correlations, also, such as the distribution of types of chromatin and how the genome is packaged in the nucleus. But I’m getting way ahead of myself.
Now, some questions. Which DNA regions of the mammalian genome are enriched in the codes for the most essential functions? Precisely where you find Alus and Alu-like sequences (the dark blue bands on the chromosome). Which sections of the mammalian genome have the highest rates of transcription? Precisely where you find Alus and Alu-like sequences. Where do you find the strongest organizational correlations between any two mammalian genomes? Precisely where you find Alus and Alu-like sequences.
Aren’t these correlations a bit strange for genomes that supposedly consist mostly of junk and are constantly being corrupted by “degenerative processes”? Why do such “obnoxious sequences” have any kind of conserved higher-order “bar code” pattern? These facts of mammalian chromosome biology have been known for years, if not decades. But, alas, no mention of them is to be found in the literature that wants to emphasize the unintelligent design of our genome. To make up for this lack, then, I am going to discuss such facts in more detail after I show you the mystery signal tomorrow.
References
1 Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521.
2 Chen TL, Manuelidis L. 1989. SINEs and LINEs cluster in distinct DNA fragments of Giemsa band size. Chromosoma 98: 309-316;
Korenberg JR, Rykowski MC. 1988. Human genome organization: Alu, lines, and the molecular structure of metaphase chromosome bands. Cell 53(3): 391-400;
Costantini M, Clay O, Federico C, Saccone S, Auletta F, Bernardi G. 2007. Human chromosomal bands: nested structure, high-definition map and molecular basis. Chromosoma 116: 29-40.
