 Evolution
Evolution
Why Proteins Aren’t Easily Recombined, Part 2
In an earlier article, I described how proteins are composed of units of secondary structure called alpha helices and beta sheets. These units of secondary structure assemble into the wide range of protein structures that exist in nature, a few of which are shown below in cartoon form. Beta strands are shown as the flattened arrows and helices as coils. Remember, proteins don’t actually look like this, but it helps us to make comparisons between protein structures when they are drawn this way.
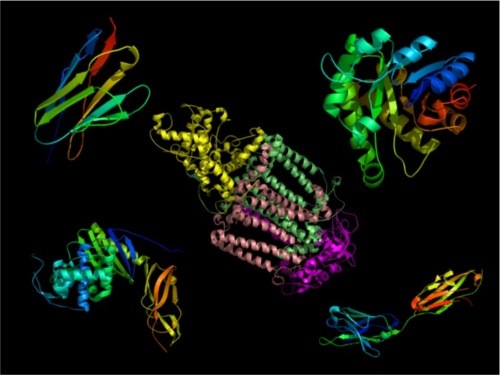
I also described why alpha helices and beta sheets (multiple beta strands grouped into sheets) are sequence-specific in their interactions. They do not function as interchangeable Lego bricks, because the exterior of each helix or strand has a unique set of side chains, with unique chemical properties.
This context-dependent behavior of structural modules, like helices and sheets, has been demonstrated experimentally. The general rule is that modules that function as independent units, with little interaction with other structural elements, can sometimes be recombined. Modules with multiple external interactions cannot. These principles are illustrated in a series of experiments using the protein beta lactamase, an enzyme that breaks down penicillin in penicillin-resistant bacteria.
First I will describe a series of experiments demonstrating that modular recombination is possible, but only in certain circumstances. Meyer and coworkers identified blocks of sequence that should function as self-contained modules in three natural beta lactamase enzymes, using a computer algorithm that analyzed the exterior surfaces of the proposed modules for side chain interactions. These enzymes had between 34-42% sequence identity to start, allowing their sequences to be aligned with some confidence. They then mixed and matched the identified self-contained modules from these enzymes into chimeric enzymes and tested them for function. Even with carefully designed splice sites, and optimal sequence independence, four out of five chimeras had no detectable function, and only one out of ten had function approaching that of the natural enzymes. Thus, even under carefully engineered, ideal circumstances, with a great deal of sequence independence, most recombinations failed.
Another test of modularity reported by Doug Axe examined whether the main structural domains of beta lactamase enzymes could be swapped. In the picture below, part (A) shows the domain structure of the two beta lactamase enzymes he used, with the two domains in each protein colored differently. In (B) the backbones of the similar domains from each protein are aligned with each other to show their 3-D similarity. Both enzymes carry out exactly the same chemical reaction, but have only 26% identical amino acid sequence.
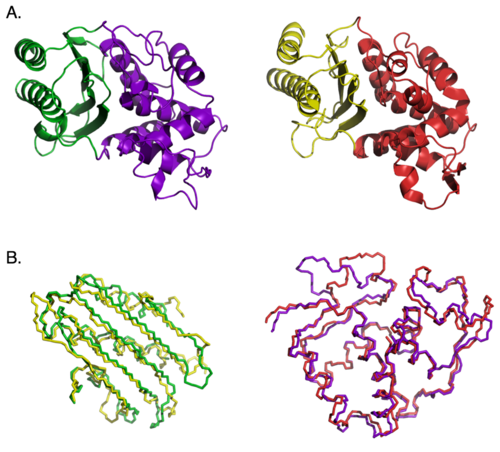
In spite of the structural and functional similarity, the domains of the two proteins are not interchangeable. Swapping domains between enzymes kills enzyme activity. Why? It’s because the active site of the enzyme lies at the interface between the two domains, and the substantially different side chain interactions at the interface disrupt the chimeric enzyme’s function. So this is a case where the extensive, specific side-chain interactions prevent any functional recombination between domains.
This same sequence specificity can be shown even at the level of individual amino acids. Using two other variants of beta lactamase whose structures are nearly identical (shown below), and whose sequences are 50% identical, another study by Axe swapped non-matching but positionally equivalent amino acids between the two proteins to see if they could substitute for one another.
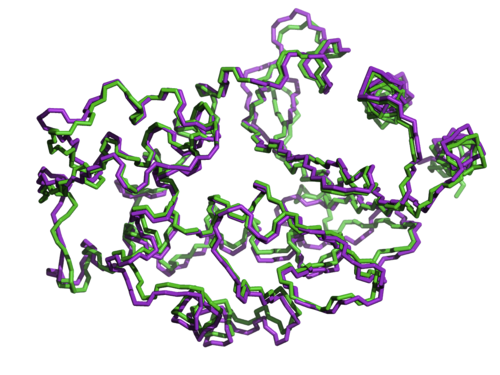
As Doug Axe described in his 2010 paper,
If aligned but non-matching residues are part for part equivalents, then we should be able to substitute freely among these equivalent pairs without impairment. Yet when protein sequences were even partially scrambled in this way, such that the hybrids were about 90% identical to one of the parents, none of them had detectable function.
In other words, even if only 10% of non-matching residues were changed, the resulting hybrid enzyme no longer functioned. Why? Because the substitution of different amino acids into the existing protein structure destabilized the fold, even though those same amino acids worked well in another context. Thus, each protein’s amino acid sequence works as a whole to help generate a proper stable fold, in a context-dependent fashion.
So we have context-dependent effects on protein function at the level of primary sequence, secondary structure, and tertiary (domain-level) structure. This does not bode well for successful, random recombination of bits of sequence into functional, stable protein folds, or even for domain-level recombinations where significant interaction is required.
Cross-posted at Biologic Perspectives.
