 Intelligent Design
Intelligent Design
 Life Sciences
Life Sciences
A Knotty Puzzle
Until fairly recently, scientists believed that folded proteins could be unfolded into a simple polypeptide chain by tugging on one end, unlike those earbuds you keep in your pocket. But it is now known that some proteins also fold themselves into knots of various kinds. This is a puzzle.
As a recent press release states,
Relatively little is known about protein folding, the process by which a polypeptide chain with a specific sequence of amino acid chains forms the three-dimensional structures — their “native states” — required to become functional.
How this process reproducibly achieves the required structure is the subject of intensive study. Even harder is understanding how this is accomplished for knotted proteins, where the chain loops around itself in entanglements of varying complexity — or the even rarer slipknotted proteins, where a loop is bound by another segment of the protein chain, similar to a shoelace bow.
What intrigued the scientists about the protein knots is that the folding process resulting in the formation of knots is intrinsically more difficult than the process producing unknotted proteins. The protein has to avoid not only energetic traps but also topological barriers.
The press release continues,
If an amino acid chain takes too much time to find its native state or if it is stuck in a misfolded or partially unfolded state, the result may be a useless protein or one that produces harm by causing protein aggregation which is known to cause neurodegenerative disorders.
“From an evolutionary point of view, knotting might seem unlikely to occur but, in fact, it does occur,” says Millett, who, along with co-first authors Joanna Sulkowska from UC San Diego and Eric J. Rawdon from the University of Saint Thomas examined, analyzed, and classified 74,223 protein structures submitted to the Protein Data Bank for the location and formation of knots.
And it appears to be a conserved feature among similar proteins. Shown below are the structures of three ubiquitin C-terminal hydrolyses from (A) human, (B) yeast, and (C) the malarial parasite P. falciparum, together with their matrix presentation. They all form the same complex knotting motif.
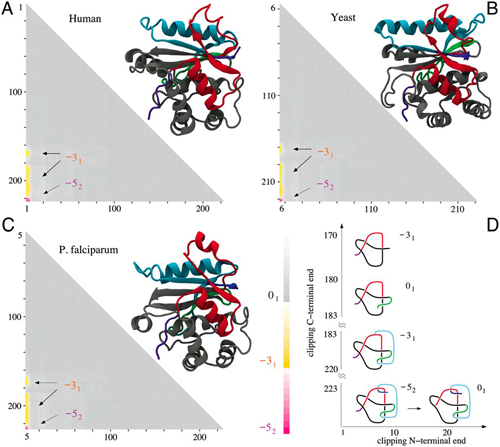
The reason for the knots remain obscure, but they may have something to do with added stability for the folded proteins, the authors state.
Let me untangle the rhetoric. The reason why knots in folded proteins are unlikely is because they are hard to achieve, without resulting in misfolded proteins, aggregation, and possible disease states. Even though it’s unlikely they evolved — let’s make that highly unlikely — we know knotted proteins must have evolved somehow, simply because they exist.
And that’s because design is knot an option.
Go here for the rest of the web article. The scientific paper is to be published in PNAS.
Cross-posted at Biologic Perspectives. Image credit: University of California, Santa Barbara.
