 Evolution
Evolution
 Intelligent Design
Intelligent Design
 Life Sciences
Life Sciences
The Humpty-Dumpty Effect: A Revolutionary Paper with Far-Reaching Implications
About two weeks ago, a Discovery Institute colleague asked me to retrieve a paper for him from the journal Protein Science. He had run across its title in a literature search, and the topic sounded interesting. As soon as I read the paper’s abstract, my pulse quickened — and then, when I read the paper itself, I immediately emailed a circle of co-workers (who share research ideas) to tell them about the paper’s tremendous significance.
Here’s why we can’t stop talking about this publication — and why you should pay attention too.
Explaining a Very Wise Observation Richard Dawkins Once Made
From time to time, Richard Dawkins says wise and insightful things. Seriously — no sarcasm intended. Among his wisest statements is the following observation from The Blind Watchmaker (italics in the original):
It is true that there are quite a number of ways of making a living — flying, swimming, swinging through the trees, and so on. But, however many ways there may be of being alive, it is certain that there are vastly more ways of being dead, or rather not alive. You may throw cells together at random, over and over again for a billion years, and not once will you get a conglomeration that flies or swims or burrows or runs, or does anything, even badly, that could remotely be construed as working to keep itself alive. (1987, p. 9)
Now, the paper I retrieved for my co-worker, entitled “The Levinthal paradox of the interactome,” Protein Science 20 (2011):2074-79, explains why the space of “being alive” is so much vastly smaller, and harder to find, than the space of being “not alive.” The paper is short (only six pages) and was written by two structural biologists, Peter Tompa of Vrije Universiteit in Brussels and George Rose of Johns Hopkins University, neither of whom is an intelligent-design advocate. But the paper’s arguments bear so strongly on the design debate, and represent so remarkable a challenge to widely held assumptions about (for instance) the origin of cells, that its effect promises to be far-reaching. As in, revolutionary.
The Levinthal Paradox, Old and New Versions
Anyone who has studied the protein folding problem will have met the famous Levinthal paradox, formulated in 1969 by the molecular biologist Cyrus Levinthal. Put simply, the Levinthal paradox states that when one calculates the number of possible topological (rotational) configurations for the amino acids in even a small (say, 100 residue) unfolded protein, random search could never find the final folded conformation of that same protein during the lifetime of the physical universe. Therefore, concluded Levinthal, given that proteins obviously do fold, they are doing so, not by random search, but by following favored pathways. The challenge of the protein folding problem is to learn what those pathways are. That’s the classical version of the paradox.
But now consider the origin of an entire cell. All cells possess what has been called an “interactome,” namely, “a complex network” comprising “a host of cellular constituents” — proteins, nucleic acids, lipids, metal ion cofactors, and so on. If the Levinthal paradox (old version) arises from the difficulty of searching the space of possible configurations for a single protein, the new version of the paradox, formulated by Tompa and Rose, asks the same question for the possible arrangements of the cell’s interactome, an enormously larger collection of objects with a correspondingly greater search space. As Tompa and Rose express the problem,
Unlike protein folding, self-assembly of the interactome has not yet prompted such widespread attention, and for understandable reasons. It is a problem of bewildering complexity…Where does one begin? Our goal here is to show that assembly of the interactome in biological real-time is analogous to folding in that the functional state is selected from a staggering number of useless or potentially deleterious alternatives.
One can grasp the heart of the problem by looking at Figure 1 (which replicates Figure 1 from the paper itself).
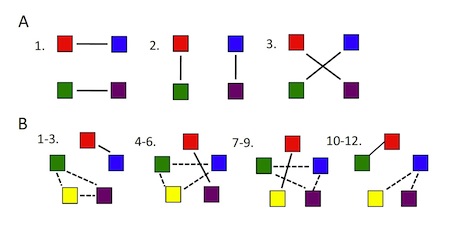
Figure 1. The number of possible cell interactomes rises much faster than the number of constituent elements.
As the number of elements — i.e., constituent proteins, illustrated here by colored squares — increases, the “number of possible different states,” or “patterns of pairwise interactions,” increases as well, but much more dramatically. With 4 proteins, there are 3 possible pairwise interactions. With 5 proteins, again counting only pairwise interactions, there are 11 possible pairwise groupings — but with 10 proteins there are 945 possible interactions (see Table 1), and with 20 proteins, 654,729,075 possible groupings. Clearly the possible interactomes for any realistic cell, with hundreds of proteins, RNAs, and other molecules, will be an enormously large number (see Table 1).
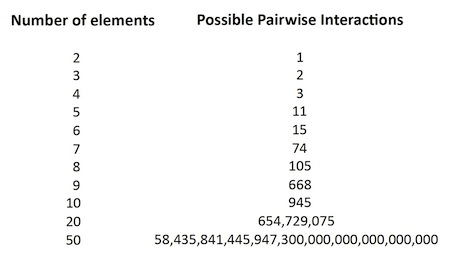
Table 1. As cellular constituents approach the total required for any actual cell, the space of possible interactomes grows beyond comprehension. Thus the “target” for the living state is exceedingly small with respect to the range of possibilities, within what Tompa and Rose call “the zone of chaos.”
Exponents Gone Wild
Tompa and Rose calculate the “total number of possible distinct patterns of interactions,” using yeast, a unicellular eukaryote, as their model system; this “total number” is the size of the space that must be searched. With approximately 4,500 proteins in yeast, the interactome search space “is on the order of 10^7200, an unimaginably large number,” they write — but “more realistic” estimates, they continue, are “yet more complicated.” Proteins present many possible surfaces for chemical interaction. “In all,” argue Tompa and Rose, “an average protein would have approximately 3540 distinguishable interfaces,” and if one uses this number for the interactome space calculation, the result is 10 followed by the exponent 7.9 x 10^10.
Tompa and Rose draw a number of lessons from their calculations. They argue, first, that any increase in biological realism will only make the Levinthal interactome paradox worse:
Of course, there are additional complicating factors such as alternative splicing, post-translational modifications, non-pairwise macromolecular interactions, incorrect complex formation that is adventitiously stable, and so forth. However, even neglecting such complications, the numbers preclude formation of a functional interactome by trial and error complex formation within any meaningful span of time. This numerical exercise…is tantamount to a proof that the cell does not organize by random collisions of its interacting constituents.
But secondly, what they call “the most profound conclusion” from their analysis bears directly on widely held assumptions about the origin of life.
The Humpty-Dumpty Effect and the Necessity for Maternal Templates
…all the king’s horses and all the king’s men / couldn’t put Humpty together again.
A highly enriched soup of proteins and nucleic acids will never form a functional cell, even if lipid bilayer membranes were provided to help these materials become organized. Indeed, the fully functional contents of a living cell, once the wall or membrane enclosing them has been breached (thus, killing the cell), move irreversibly in the direction of non-living chemistry. Humpty Dumpty, once he cracks, does not reconstitute, but enters what Tompa and Rose call the “zone of chaos,” never to return.
Tompa and Rose have sketched the theoretical basis for why this happens:
[O]ur calculations of combinatorial complexity [show] that the emergent interactome could not have self-organized spontaneously from its isolated protein components. Rather, it attains its functional state by templating the interactome of a mother cell and maintains that state by a continuous expenditure of energy. In the absence of a prior framework of existing interactions, it is far more likely that combined cellular constituents would end up in a non-functional, aggregated state, one incompatible with life…The spontaneous origination of a de novo cell has yet to be observed; all extant cells are generated by the division of pre-existing cells that provide the necessary template for perpetuation of the interactome.
Tompa and Rose spell out other implications of their analysis (e.g., for medicine and synthetic biology), but maybe we’ve piqued your curiosity enough already. This paper deserves your attention. As noted, for a close circle of us at Discovery and Biologic, it’s the most interesting and significant paper we’ve read in years.
Image: Denslow’s Humpty Dumpty/Wikicommons.

