 Evolution
Evolution
 Life Sciences
Life Sciences
Gene Duplication and the Origin of Novel Biological Information: A Case Study of the Globins
Those of us who have been involved with the debate surrounding intelligent design and evolution for any significant length of time are well acquainted by now with the most fashionable neo-Darwinian model for the origin of novel biological information: gene duplication and divergence. Gene duplications normally arise through a phenomenon known as “unequal cross-over,” which occurs during cell division. This process results in the deletion of a sequence in one strand, and its replacement with a duplicate from its homologous chromosome (meiosis) or its sister chromatid (mitosis). The model of gene duplication and divergence essentially maintains that, following a gene duplication, while one copy of the gene retains its original function, the other copy is freed from selective constraint and is thus free to mutate at a faster rate, and explore sequence space in search of some novel function.
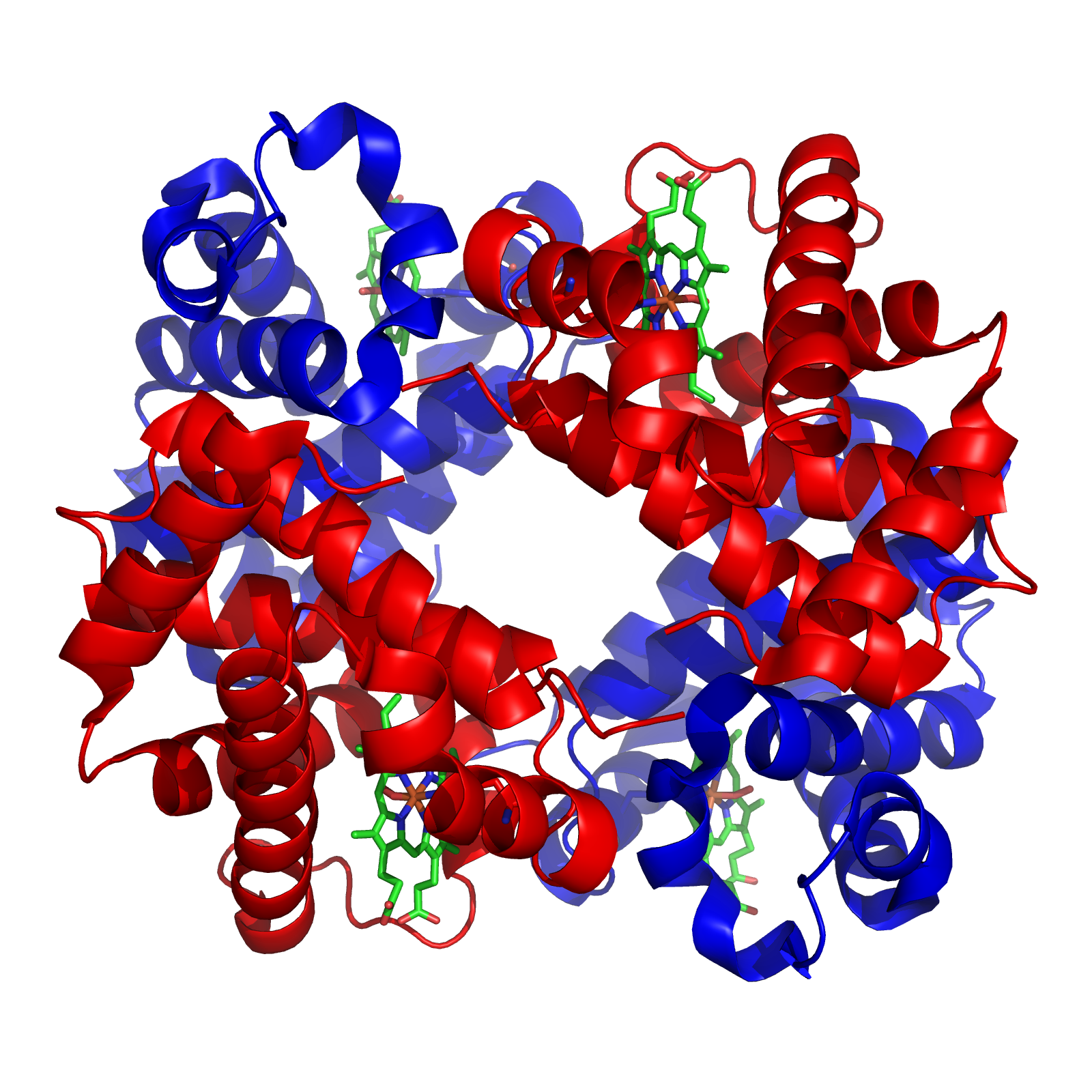
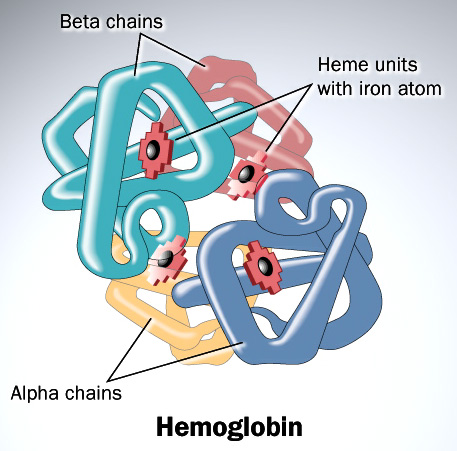
With this background, I wish to conduct a short case study of the evolution of the globin family: heme-containing proteins that are characterized by their incorporation of the globin fold (a series of eight alpha helical segments). The globins are involved in the binding and/or transport of oxygen, the two best-known examples being the reversible oxygen-binders hemoglobin (featured in the diagram above) and myoglobin.
As many readers of ENV will be well aware, the sequence of amino acids in a polypeptide chain determines its uniquely folded three-dimensional conformation, which is largely determined by the distribution of polar and non-polar side chains. Proteins typically possess a shell of hydrophilic amino acid residues that surround an inside-core of hydrophobic residues, which have been buried so as to avoid contact with water. The peptide bonds hydrogen-bond with each other to give a so-called “secondary structure,” classified as α-helices and β-sheets. A structure of multiple adjacent elements of secondary structure is known as a supersecondary structure or motif, and includes α-helix hairpins, β-hairpins, and β-α-β motifs. When different motifs pack together they form domains, which comprise the fundamental units of a protein’s tertiary structure. When several polypeptide chains associate into an oligomeric molecule, it is referred to as the protein’s quaternary structure. For example, the protein hemoglobin is a tetramer comprised of two structurally identical α chains and two structurally identical β chains (these chains are also referred to as “subunits”). In the diagram of the structure of hemoglobin (above), the different chains are colored red and blue, and the iron-containing heme groups are colored green.
Hemoglobin is responsible for carrying oxygen from the lungs (or gills in the case of fish) to the tissues of the body where the oxygen is released to provide energy for the organism. Hemoglobin also serves to collect carbon dioxide to bring it back to the lungs so that it can be expelled from the organism’s body.
The Adaptive Value of Proposed Intermediates
 Consider the divergence between the α and β chains of hemoglobin. Following the duplication of the initial hemoglobin gene, each copy has to diverge simultaneously, and in complementary ways in order to ensure a functional tetramer. For one thing, characteristic of hemoglobin is that both chains possess hydrophobic residues which are essential for the association of the subunits (this stands in marked contrast to myoglobin which, being a water-soluble protein, possesses mostly hydrophilic residues on the outside of its folded structure).
Consider the divergence between the α and β chains of hemoglobin. Following the duplication of the initial hemoglobin gene, each copy has to diverge simultaneously, and in complementary ways in order to ensure a functional tetramer. For one thing, characteristic of hemoglobin is that both chains possess hydrophobic residues which are essential for the association of the subunits (this stands in marked contrast to myoglobin which, being a water-soluble protein, possesses mostly hydrophilic residues on the outside of its folded structure).
It is conventional wisdom that the early hemoglobin may have been a monomer (similar to myoglobin), which possessed exterior hydrophilic residues, some of which were subsequently substituted for hydrophobic residues. For example, in their book, Hemoglobin: Structure, Function and Evolution (1983), Richard Dickerson and Irving Geis argue that some of these substitutions occurred prior to the gene duplication: That is to say, the early hemoglobin evolved into a tetramer (or possibly a dimer) prior to its diverging into the α and β chains. Such a suggestion is, however, suspect in light of the fact that one might expect to see these hydrophobic amino acids in similar positions if they arose prior to the divergence of the hemoglobins. But the hydrophobic amino acids on the exterior of the polypeptides are different and are located in different positions on the α and β chains. This promotes their complementarity: Hydrophobicity is only needed at the sites that associate with the other chains (not necessarily at the same location on the different chains).
The β chains, unlike the α chains, are unable to make contact with one another. When the β chains are not produced (as in β-thalassemia, a condition that is often fatal), the unassociated α chains typically degrade. In the case of α-thalassemia, however, where it is the α chains that are lacking, the β chains do associate. This presents a different difficulty, for they bind oxygen with an affinity similar to that of myoglobin. This means that the oxygen will not be released to the tissues — again, with fatal consequences.
Given the plethora of pathological disorders that may be attributed to a change in just one amino acid in these chains (thus disrupting chain cooperativity), it should be evident that many amino acids in both chains are essential for the correct functioning of tetrameric hemoglobin.
Thus, as I hope to have shown above for our current case study (i.e. the globins), for gene family trees to be considered credible, it is necessary to show that there is a sufficiently high likelihood of all of the ancestral sequences conferring some sort of selective advantage. But it has been shown time and again, as I discuss here, that getting from one set of conserved amino acids to another — as is necessary for the production by divergence of proteins with different functions — is too big a jump through sequence space.
The Fragility Problem
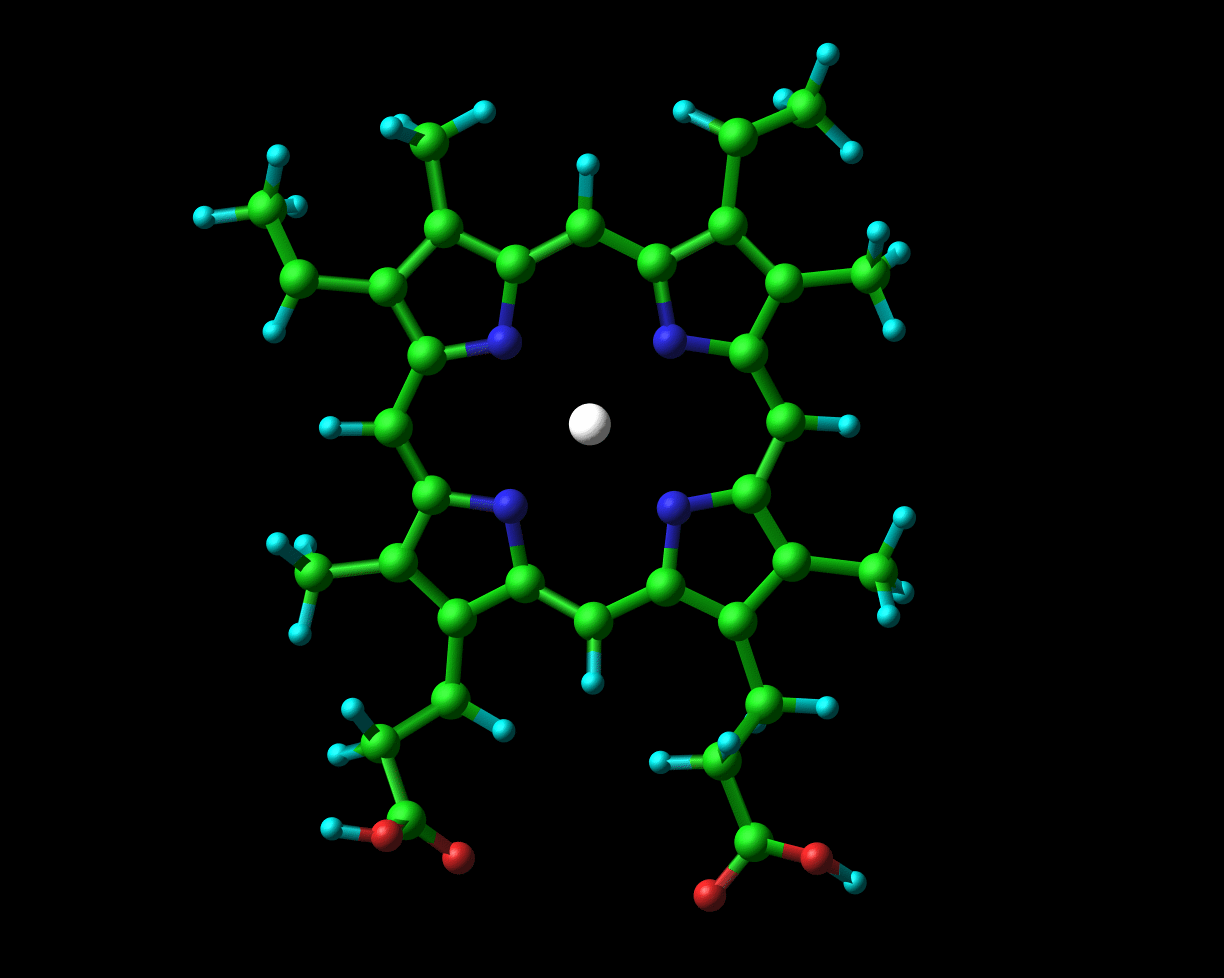
Globins possess a heme group (pictured on the left) which contains iron. Iron exists in two oxidized forms, namely Fe2+ and Fe3+ (ferrous and ferric forms respectively). When oxygen is available, the iron is readily oxidized to ferric Fe3+. But, crucial to its physiological role, the iron that is present in hemoglobin and myoglobin exists in the ferrous oxidation state. In Chapter 2 of their book to which I previously alluded, Dickerson and Geis observe that “The purpose of the heme and the polypeptide chain around it is to keep the ferrous iron from being oxidized (metmyoglobin, with a ferric iron, does not bind oxygen), and to provide a pocket into which the oxygen can fit.”
The ferrous iron, in hemoglobin and myoglobin, is actively protected from being oxidized to the ferric form by the specific chemical groups of the surrounding amino acid residues. Dickerson and Geis describe the fragility of this system as follows:
The met [oxidized] forms were experimentally the easiest to obtain, and the deoxy states also could be crystallized and studied with careful experimental techniques. The oxy forms proved more intractable: Unless extreme care is taken, O2 oxidizes the heme iron from Fe2+ to Fe3+ rather than simply binding, thus yielding the unwanted met form of the molecule.
As one might expect, the amino acids that surround the heme group are evolutionarily highly conserved. Moreover, many pathological conditions arise as the result of changing just one amino acid, resulting in the inability of the polypeptide to retain the heme group correctly, thus permitting the iron to oxidize. In many cases, changing just one amino acid alters the positioning of the amino acids next to the heme group, such that they are no longer able to protect it from oxidation.
Complementary Changes Involving the Regulation of Gene Expression
It often seems that evolutionary scientists are so interested in the similarity of structure and function of the proteins hemoglobin and myoglobin that they completely neglect the fact that they are produced in totally different tissues: bone marrow and muscle respectively. Myoglobin’s function is made possible by its higher affinity for oxygen. The modifications of the protein’s amino acid sequence, in order for it to be converted from hemoglobin to myoglobin, would have needed to be accompanied by complementary changes in its regulatory sequences in order to ensure that the myoglobin was produced in the muscle where it is needed, rather than in bone marrow where the red blood cells are produced. Myoglobin present in red blood cells would not provide a selective advantage. In fact, it would be harmful to the organism because it would bind too tightly to oxygen and not release it to the tissues (as in the case of α-thalassemia described above). This is one of the most neglected points in discussions concerning gene duplication and family trees.
Conclusion
In summary, we have seen that the scope for evolution of novel genes and proteins by virtue of gene duplication and subsequent divergence or recruitment is very limited, even in facilitating relatively trivial functional innovations. Given the extremely diverse array of protein conformations found in living systems, the likelihood of the relatedness of genes — even within gene families — may be treated with suspicion and healthy skepticism. It is somewhat ironic that biologists are all too willing to accept a statistical argument against two or more proteins with similar sequences arising independently by chance, but are completely unwilling to consider statistical arguments against them arising by chance at all.
