 Intelligent Design
Intelligent Design
The Molecular “Clutch” of the Dynein Motor Protein

Here at ENV, I have previously described the molecular flagellar clutch of Bacillus subtilis, the grass or hay bacillus, which allows the bacterium to cease motility upon biofilm formation. A new paper, published in the journal Cell, reports on the discovery of a similar clutch associated with the motor protein dynein.
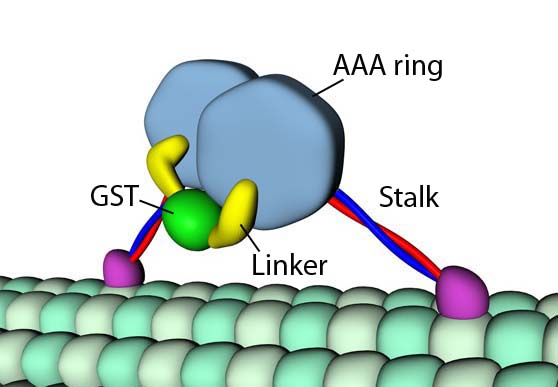 Dynein is similar to kinesin inasmuch as it transports molecular cargo along microtubule “railways.” Unlike kinesin, which walks towards the “plus” end of the microtubule, dynein walks in the direction of the “minus” end. There are also important structural differences. For example, dynein possesses a ring-shaped ATPase domain (the dynein head) comprised of six AAA+ modules and a seventh non-AAA+ domain (Roberts et al., 2009).
Dynein is similar to kinesin inasmuch as it transports molecular cargo along microtubule “railways.” Unlike kinesin, which walks towards the “plus” end of the microtubule, dynein walks in the direction of the “minus” end. There are also important structural differences. For example, dynein possesses a ring-shaped ATPase domain (the dynein head) comprised of six AAA+ modules and a seventh non-AAA+ domain (Roberts et al., 2009).
The functions of dynein include “transporting and positioning diverse cargos (e.g., mRNAs, proteins, and organelles) during interphase, exerting tension between the microtubule network and cell cortex during cell migration, and helping to construct the spindle during mitosis and meiosis” (Huang et al., 2012).
Although dynein is somewhat cumbersome relative to kinesin, this increased structural complexity provides the cell with the capacity to dynamically regulate its activity, literally shifting gears in response to the load, and regulating its speed in response to the cell’s needs (Mallik and Gross, 2004). Indeed, defects in the regulation of dynein can result in a number of human diseases (Gerdes and Katsanas, 2005).
But how exactly does this motor protein regulate its speed and activity? How is it able to halt its progression along the microtubule rails when the cargo has arrived at its target location? The lissencephaly protein (Lis1) is known to be responsible for regulating dynein’s activity (Mesngon et al., 2006; Tai et al., 2002; Sasaki et al., 2000; Reiner et al., 1993). In fact, “mutations in Lis1 cause classical lissenchephaly, a developmental brain abnormality characterized by defects in neuronal positioning,” (Mesngon et al., 2006). The precise mechanism whereby Lis1 regulates the activity of dynein, however, is less well characterized.
The new paper in Cell reports:
The lissencephaly protein Lis1 has been reported to regulate the mechanical behavior of cytoplasmic dynein, the primary minus-end-directed microtubule motor. However, the regulatory mechanism remains poorly understood. Here, we address this issue using purified proteins from Saccharomyces cerevisiae and a combination of techniques, including single-molecule imaging and single-particle electron microscopy. We show that rather than binding to the main ATPase site within dynein’s AAA+ ring or its microtubule-binding stalk directly, Lis1 engages the interface between these elements. Lis1 causes individual dynein motors to remain attached to microtubules for extended periods, even during cycles of ATP hydrolysis that would canonically induce detachment. Thus, Lis1 operates like a “clutch” that prevents dynein’s ATPase domain from transmitting a detachment signal to its track-binding domain.
To visualize what was going on, the researchers used “purified proteins from Saccharomyces cerevisiae and a combination of techniques, including single-molecule imaging and single-particle electron microscopy.”
The paper further reports,
Here, using functional, recombinant proteins from S. cerevisiae, we show that Lis1 engages dynein at AAA3/AAA4. From this site, Lis1 acts like a “clutch” to regulate communication between dynein’s catalytic ring and microtubule-binding stalk, promoting a microtubule-bound state. Supporting these results, we identify mutations at the AAA3/4 junction that drastically impair Lis1 binding and motility regulation in vitro and dynein function in vivo. In addition, we identify an arginine finger motif within AAA4 and find that its mutation mimics aspects of Lis1’s effect in vitro. Previous genetic studies in an evolutionarily distant filamentous fungus showed that the same mutation can partially rescue Lis1 loss in vivo (Zhuang et al., 2007). These results allow us to propose how Lis1 biases dynein to a microtubule attached-state and assists in a variety of cellular functions across eukaryotes. [emphasis added]
If you drive an automobile with a manual transmission, the clutch pedal is of course the one to the left of the brake and the gas. Just as a clutch disengages a car engine from the transmission system, the Lis1 protein disengages the dynein motor from its walking appendages, by attaching where these appendages would connect to the motor.
Come to think of it, this all sounds remarkably like what an engineer would do. Oh…wait.
Image: 2011 Citroen DS3, Flickr/NRMA New Cars.
