 Evolution
Evolution
 Intelligent Design
Intelligent Design
Exon Shuffling, and the Origins of Protein Folds
A frequently made claim in the scientific literature is that protein domains can be readily recombined to form novel folds. In Darwin’s Doubt, Stephen Meyer addresses this subject in detail (see Chapter 11). Over the course of this and a subsequent article, I want to briefly expand on what was said there.
Defining Our Terms
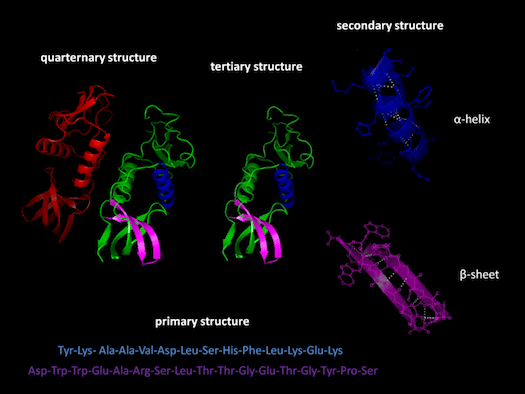
Before going on, it may be useful for me to define certain key terms and concepts. I will be referring frequently to “exons” and “introns.” Exons are sections of genes that code for proteins; whereas introns are sections of genes that don’t code for proteins.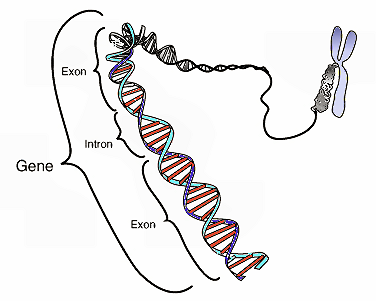 Proteins have multiple structural levels. Primary structure refers to the linear sequence of amino acids comprising the protein chain. When segments within this chain fold into structures such as helices and loops, this is referred to as secondary structure. Common units of secondary structure include α-helices and β-strands. Tertiary structure is the biologically active form of the protein, and refers to the packing of secondary structural elements into domains. Since a protein’s tertiary structure optimizes the forces of attraction between amino acids, it is the most stable form of the protein. When multiple folded domains are arranged in a multi-subunit complex, it is referred to as a quaternary structure.
Proteins have multiple structural levels. Primary structure refers to the linear sequence of amino acids comprising the protein chain. When segments within this chain fold into structures such as helices and loops, this is referred to as secondary structure. Common units of secondary structure include α-helices and β-strands. Tertiary structure is the biologically active form of the protein, and refers to the packing of secondary structural elements into domains. Since a protein’s tertiary structure optimizes the forces of attraction between amino acids, it is the most stable form of the protein. When multiple folded domains are arranged in a multi-subunit complex, it is referred to as a quaternary structure.
A further concept is domain shuffling. This is the hypothesis that fundamentally new protein folds can be created by recombining already-existing domains. This is thought to be accomplished by moving exons from one part of the genome to another (exon shuffling). There are various ways in which exon shuffling might be achieved, and it is to this subject that I now turn.
The Mechanisms of Exon Shuffling
There are several ways in which exon shuffling may occur. Exon shuffling can be transposon-mediated, or it can occur as a result of crossover during meiosis and recombination between non-homologous or (less frequently) short homologous DNA sequences. Alternative splicing is also thought to play a role in facilitating exon shuffling.
When domain shuffling occurs as a result of crossover during sexual recombination, it is hypothesized that it takes place in three stages (called the “modularization hypothesis”). First, introns are gained at positions that correspond to domain boundaries, forming a “protomodule.” Introns are typically longer than exons, and thus the majority of crossover events take place in the noncoding regions. Second, within the inserted introns, the newly formed protomodule undergoes tandem duplication. Third, intronic recombination facilitates the movement of the protomodule to a different, non-homologous, gene.
Another hypothesized mechanism for domain shuffling involves transposable elements such as LINE-1 retroelements and Helitron transposons, as well as LTR retroelements. LINE-1 elements are transcribed into an mRNA that specifies proteins called ORF1 and ORF2, both of which are essential for the process of transposition. LINE-1 frequently associates with 3′ flanking DNA, transporting the flanking sequence to a new locus somewhere else on the genome (Ejima and Yang, 2003; Moran et al., 1999; Eickbush, 1999). This association can happen if the weak polyadenylation signal of the LINE-1 element is bypassed during transcription, causing downstream exons to be included on the RNA transcript. Since LINE-1’s are “copy-and-paste” elements (i.e. they transpose via an RNA intermediate), the donor sequence remains unaltered.
Long-terminal repeat (LTR) retrotransposons have also been established to facilitate exon shuffling, notably in rice (e.g. Zhang et al., 2013; Wang et al., 2006). LTR retrotransposons possess a gag and a pol gene. The pol gene translates into a polyprotein composed of an aspartic protease (which cleaves the polyprotein), and various other enzymes including reverse transcriptase (which reverse transcribes RNA into DNA), integrase (used for integrating the element into the host genome), and Rnase H (which serves to degrade the RNA strand of the RNA-DNA hybrid, resulting in single-stranded DNA). Like LINE-1 elements, LTR retrotransposons transpose in a “copy-and-paste” fashion via an RNA intermediate. There are a number of subfamilies of LTR retrotransposons, including endogenous retroviruses, Bel/Pao, Ty1/copia, and Ty3/gypsy.
Alternative splicing by exon skipping is also believed to play a role in exon shuffling (Keren et al., 2010). Alternative splicing allows the exons of a pre-mRNA transcript to be spliced into a number of different isoforms to produce multiple proteins from the same transcript. This is facilitated by the joining of a 5′ donor site of one intron to the 3′ site of another intron downstream, resulting in the “skipping” of exons that lie in between. This process may result in introns flanking exons. If this genomic structure is reinserted somewhere else in the genome, the result is exon shuffling.
There are of course other mechanisms that are hypothesized to play a role in exon shuffling. But this will suffice for our present purposes. Next, we will look at the evidence for and against domain shuffling as an explanation for the origin of new protein folds.
Introns Early vs. Introns Late
It was hypothesized fairly early, after the discovery of introns in vertebrate genes, that they could have contributed to the evolution of proteins. In a 1978 article in Nature, Walter Gilbert first proposed that exons could be independently assorted by recombination within introns (Gilbert, 1978). Gilbert also hypothesized that introns are in fact relics of the original RNA world (Gilbert, 1986). According to the “exons early” hypothesis, all protein-coding genes were created from exon modules — coding for secondary structural elements (such as α-helices, β-sheets, signal peptides, or transmembrane helices) or folding domains — by a process of intron-mediated recombination (Gilbert and Glynias, 1993; Dorit et al., 1990).
The alternative “introns late” scenario proposed that introns only appeared much later in the genes of eukaryotes (Hickey and Benkel, 1986; Sharp, 1985; Cavalier-Smith, 1985; Orgel and Crick, 1980). Such a scenario renders exon shuffling moot in accounting for the origins of the most ancient proteins.
The “introns early” hypothesis was the dominant view in the 1980s. The frequently cited evidence for this was the then widespread belief in the general correspondence between exon-intron structure and protein secondary structure.
From the mid 1980s, this view became increasingly untenable, however, as new information came to light (e.g. see Palmer and Logsdon, 1991; and Patthy, 1996; 1994; 1991; 1987) that raised doubts about a general correlation between protein structure and intron-exon structure. Such a correspondence is not borne out in many ancient protein-coding genes. Moreover, the apparently clearest examples of exon shuffling all took place fairly late in the evolution of eukaryotes, becoming significant only at the time of the emergence of the first multicellular animals (Patthy, 1996; 1994).
In addition, analysis of intron splicing junctions suggested a similar pattern of late-arising exon shuffling. The location where introns are inserted and interrupt the protein’s reading frame determines whether exons can be recombined, duplicated or deleted by intronic recombination without altering the downstream reading frame of the modified protein (Patthy, 1987). Introns can be grouped according to three “phases”: Phase 0 introns insert between two consecutive codons; phase 1 introns insert between the first and second nucleotide of a codon; and phase 2 introns insert between the second and third nucleotide.
Thus, if exon shuffling played a major role in protein evolution, we should expect a characteristic intron phase distribution. But the hypothetical modules of ancient proteins do not conform to such expectations (Patthy, 1991; 1987).
Conclusion
It is clear, then, that exon shuffling (at the very least) is unlikely to explain the origins of the most ancient proteins that have emerged in the history of life. But is this mechanism adequate to explain the origins of later proteins such as those that arise in the evolution of eukaryotes? Tomorrow I will evaluate the evidence pro-and-con for the role of exon shuffling in protein origins.
