 Evolution
Evolution
 Intelligent Design
Intelligent Design
 Life Sciences
Life Sciences
Molecular Machines Reach Perfection
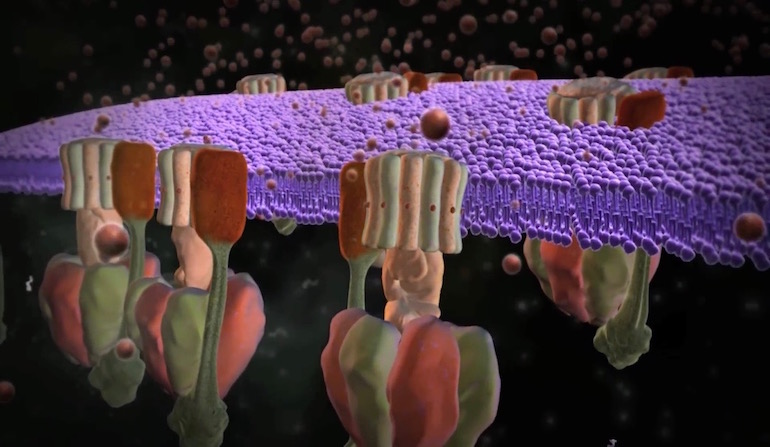
ATP synthase is in the news again, and it’s even better than before. Before hearing the news, it might be worthwhile to review our animation of this tiny rotary engine that powers all life, from bacteria to humans. You’re running on quadrillions of these little motors right now. The news is that they are perfect.
One doesn’t often see the word “perfect” in a science paper, but four Japanese researchers are unabashed, using the word 13 times in their paper in the Proceedings of the National Academy of Sciences, including the title: “Perfect chemomechanical coupling of F0F1-ATP synthase.”
Peter D. Mitchell, a Nobel awardee in 1978, proposed that F0F1-ATP synthase converts energy between electrochemical potential of H+ across biological membrane…, which is established by respiratory chain complexes, and chemical potential of adenine nucleotide [ΔG(ATP)]. However, the efficiency of the energy conversion has been a matter of debate for over 50 years. In this study, with a highly reproducible analytical system using F0F1-ATP synthase from thermophilic Bacillus, apparently perfect energy conversion was observed. Mitchell’s prediction thus has quantitative evidence. [Emphasis added.]
You can’t get better than perfect. This means that every proton (H+) coming into the machine, driving its rotation, yields 100 percent conversion of its energy into production of ATP. Can you think of any man-made motor that even approaches this kind of efficiency? Hardly. At our macro level of engineering, artificial motors waste energy through heat, friction, and escape of fuel to the environment. The second law of thermodynamics forbids perfection. Somehow, at the scale of nanometers, ATP synthase engines get maximum bang for their proton buck — with no loss at all.
The debate about ATP synthase energy efficiency centered on the numerical mismatch between the two halves of the machine. The F0 part, where protons enter, has 10 units called c subunits arranged like orange peels that rotate around a central axis. The F1 part, by contrast, has 3 units in pairs, called β subunits, where ATP synthesis takes place (the two halves are linked by a central stalk called the γ-subunit that works like a camshaft). This 10/3 non-integer pairing between F0 and F1 was unexpected, leading biophysicists to assume there must be some slippage in the camshaft during every rotation. Slippage would waste some of the proton motive force (pmf), reducing the efficiency.
One way to find the answer is to compare the input to the output as accurately as possible. These scientists rigged a proteoliposome from a thermophilic (heat-loving) bacterium in a new way that allowed them to reliably measure the incoming pmf as well as the outgoing production of ATP.
In this report, we used this system to determine the actual H+/ATP ratio. The results show the perfect agreement of H+/ATP ratio to c/β, indicating tight coupling efficiency of proton translocation in Fo and ATP synthesis/hydrolysis in F1. In addition, kinetic and energetic equivalence of transmembrane difference of pH (ΔpH) and electric potential (Δψ) was supported with unprecedented certainty over a wide range of pmf values.
The team carefully eliminated all contamination, ran the tests for tens of hours, and reduced error to achieve unprecedented levels of accuracy. “A long-anticipated, but unproved, conception that F0F1 achieves a perfect coupling between transmembrane H+ translocation and ATP synthesis/hydrolysis has direct experimental evidence now,” they conclude.
How is this even possible? Isn’t there slippage? Isn’t there twisting force of torque as the camshaft presses against the β subunits in F1? And what about other versions of ATP synthase in other organisms that have 8, 12, or 14 c subunits in F0? They address these questions in the final paragraph of the Discussion:
In a thermodynamic view, the perfect coupling means perfect energy conversions between chemiosmotic (H+ translocation), mechanical (rotary motion), and chemical energy (ATP synthesis/hydrolysis). A near-perfect energy conversion from ATP hydrolysis to rotary motion of γ-subunit in F1 was recently demonstrated in a thermodynamically defined manner, and this study predicts that other conversions should also be highly efficient. In a mechanistic view, the perfect coupling means that there is no slippage within and between F0 motor and F1 motor. Atomic structures of F1 are convincing that rotary motion of the γ-subunit could not occur without conformation change of the catalytic subunits. Structural basis for rotation of F0 motor without slippage has been suggested recently by atomic structures of whole F0F1 revealed by cryoelectron microscopy. The connection of the two motors should also be strong enough to endure the twisting force of torque. Crystal structures of F1·c-ring complexes indicate that the connection appears to be held by a small number of interactions between the bottom portion of F1’s rotor and polar loops in the c ring. Interestingly, this connection must be versatile, because the chimera TF0F1 with replaced F0 from Propionegenium modestum that has 11 c subunits shows good coupled activity.
This is a remarkable thing. Perfect — yet versatile! You can substitute a different c-ring into F0 and still get “good coupled activity.” Try that with man-made engines!
Other Perfect Scores
Kinesin, the walking machine (see our animation), is another “perfect 10” performer. Like ATP synthase, it converts chemical energy into mechanical energy. It even has what scientists call a “power stroke” as it walks. Tomonami Sumi from Okayama University in Japan compared the machine’s walking efficiency to its ATP consumption. Publishing in Nature Scientific Reports, he found that “the ratio of the number of ATP hydrolysis to the number of steps advanced suggests a tight coupling between the two.” Tight coupling; we heard that in the previous story. Although he doesn’t use the word perfect, he speaks admiringly of the “extraordinary motor properties” of kinesin. It appears that the Japanese are less inhibited about using the d-word design. Sumi’s title is, “Design principles governing chemomechanical coupling of kinesin.”
Cohesin and condensin are proteins that help keep DNA organized. An interesting article written like a mystery story in Nature News shows how scientists are trying to figure out if they work like motors. Writer Elie Dolgin calls it “DNA’s secret weapon against knots and tangles.” Something is seen extruding loops in DNA, working to “keep local regions of DNA together, disentangling them from other parts of the genome and even giving shape and structure to the chromosomes.” But whatever it is, it has to be beyond belief if MIT biophysicist Leonid Mirny’s model is correct:
For one thing, the identity of the molecular machine that forms the loops remains a mystery. If the leading protein candidate acted like a motor, as Mirny proposes, it would guzzle energy faster than it has ever been seen to do. “As a physicist friend of mine tells me, ‘This is kind of the Higgs boson of your field’,” says Mirny; it explains one of the deepest mysteries of genome biology, but could take years to prove.
The race is on to discover what kind of motor is consuming ATP to push and pull DNA. Loop extrusion not only prevents knots and tangles, it regulates gene expression by keeping parts of genes in proximity. We expect this mystery will have a “perfect” ending.
Lastly, that familiar icon the bacterial flagellum shows a new trick up its sleeve. How does the driveshaft know when to stop growing? A paper in Science shows that the “most efficient machine in the universe,” as Howard Berg calls it, has a perfect solution: it grows till it touches the periplasm (outer membrane layer). As we marvel at the engineering, let’s give evolution the credit, shall we?
The bacterial flagellum exemplifies a system where even small deviations from the highly regulated flagellar assembly process can abolish motility and cause negative physiological outcomes. Consequently, bacteria have evolved elegant and robust regulatory mechanisms to ensure that flagellar morphogenesis follows a defined path, with each component self-assembling to predetermined dimensions. The flagellar rod acts as a driveshaft to transmit torque from the cytoplasmic rotor to the external filament. The rod self-assembles to a defined length of ~25 nanometers. Here, we provide evidence that rod length is limited by the width of the periplasmic space between the inner and outer membranes. The length of Braun’s lipoprotein determines periplasmic width by tethering the outer membrane to the peptidoglycan layer.
Science Daily adds, “To function properly and propel the bacterium, the flagellum requires all of its components to fit together to exacting measurements.” The growing “driveshaft” somehow feels the outer layer and knows to stop growing. “The rod needs to touch the inside of the outer membrane,” one of the authors says. “So, if the outer membrane is farther away, the rod has to grow there to meet it.” The versatile growth process yields a perfect fit.
If you can think of any machine in your experience that is perfect yet flexible, it probably did not come about through blind, aimless natural processes. Let’s stop allowing Darwinians to get away, unchallenged, with saying they “have evolved” to perfection.
Image: ATP synthase, from “ATP Synthase: The Power Plant of the Cell.”
