 Evolution
Evolution
 Intelligent Design
Intelligent Design
New BIO-Complexity Paper Details Complexity of Function and Assembly of Bacterial Flagellum
In July I covered a paper in the journal BIO-Complexity that reviewed engineering constraints on the bacterial flagellum. I noted that the author, Waldean Schulz, an engineer with a PhD in computer science, was preparing future papers that would explore questions about flagellar evolution, such as, “Would such partial systems be preserved long enough for additional cooperating components to evolve?” Now Schulz has published a second peer-reviewed scientific paper in BIO-Complexity, “An Engineering Perspective on the Bacterial Flagellum: Part 2 — Analytic View,” which begins to address that question.
A “Bottom Up” Approach
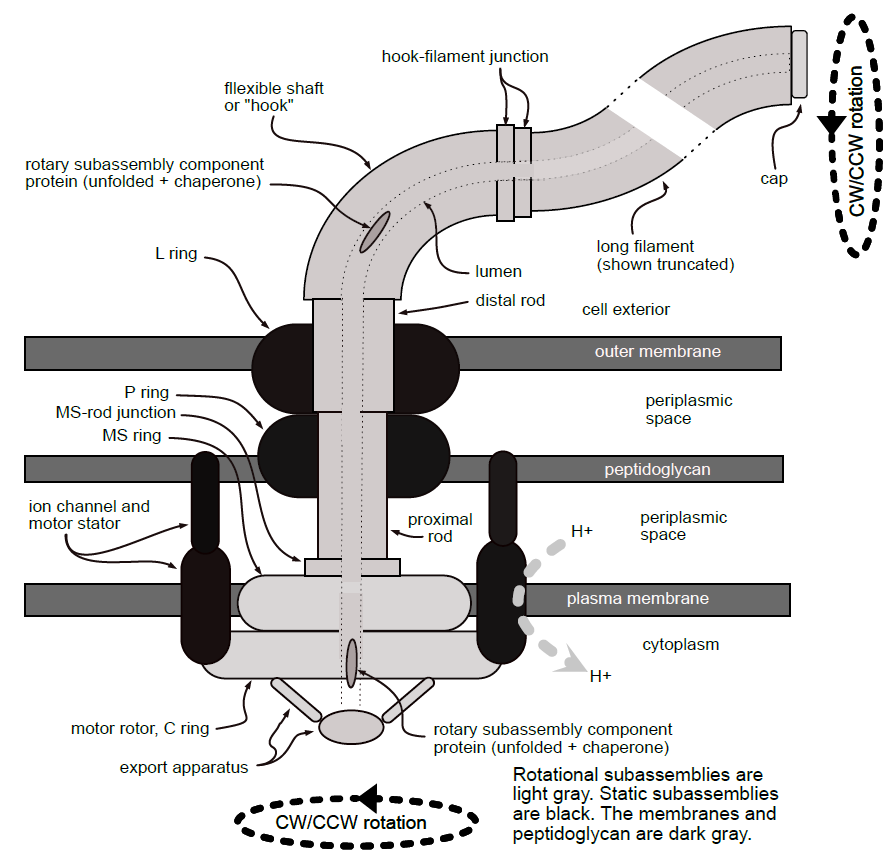
His first paper took a “top down” approach to determining the components and design necessary to fulfill the functional constraints of the flagellum. The second paper takes a “bottom up” approach, and reviews “the known 40+ protein components and the observed and inferred structure, control, and assembly of a typical bacterial flagellum.” He provides a clear diagram of an “archetypical flagellum,” labeling features such as the motor stator, filament, hook, and rod:
Schulz notes that the assembly process requires a regular set of components and procedures in order to work properly — on this point, he quotes Cohen et al. (2017) from Science who state:
The bacterial flagellum exemplifies a system where even small deviations from the highly regulated flagellar assembly process can abolish motility and cause negative physiological outcomes. Consequently, bacteria … [possess] robust regulatory mechanisms to ensure that flagellar morphogenesis follows a defined path, with each component self-assembling to predetermined dimensions.
Schulz further explains that these protein components themselves are fine-tuned such that “all proteins in each rotary subassembly need to (non-covalently) bind tightly to themselves and to the proteins of the adjacent subassemblies. This is noteworthy: the combinatorial configurations of the ensemble of proteins must be very specifically orchestrated.”
Biochemical Basis of Chemotaxis
Schulz considers not just the design and construction of the flagellum, but also the biochemical basis of chemotaxis. It’s a long passage but it’s worth reading to appreciate the complexity, elegance, and control of the system:
While there are diverse chemotaxis systems for differing bacteria, generally the concentrations of environmental chemistry are sensed by transmembrane methyl-accepting chemotaxis proteins (MCP). In the case of our archetypical bacterium, these MCPs include Tar, Tsar, Trg, Tap (E. coli) or Tcp (Salmonella), and Aer. They have a periplasmic ligand binding region and a cytosolic signalling region. These are bound to CheA by CheW to form clusters located at one or both polar ends. CheA auto-phosphorylates according to the methylation of the MCPs. Presence of nutrients increases methylation; toxins or repellants decrease it. The methylation state implements the short-term memory of the sensor system. Rebbapragada states, “Repellent binding to a chemotaxis receptor induces a conformational change in the signalling domain [of the MCP] that increases the rate of CheA autophosphorylation. The phosphoryl residue from CheA is transferred to CheY.” CheY-P in its phosphorylated state, diffuses to a flagellum, and binds to the flagellar rotor. That causes the rotor to switch the direction of rotation of the filament so that the bacterium tumbles. Tumbling causes a random new direction for forward travel after CheZ dephosphorylates CheY-P. Then CheY unbinds from the switch, default (counterclockwise) rotation ensues, and forward travel resumes. This periodic tumbling occurs about every second. The upshot is a biased random “climb” up an attractant gradient.
Meanwhile the methyl esterase protein CheB demethylates the MCP-Che-CheW complex, eventually resetting it back to its non-signalling state. The response of CheB is slower than the transfer of phosphorylation to CheY, so CheY-P can interact with the flagellar motor rotor before CheW responds (as a kind of delayed negative feedback). In parallel, CheZ removes the phosphorylated state of CheY. So, CheB and CheZ provide adaptation (hysteresis) by a time-delayed negative feedback. If stimuli are present in abundance, the phosphorylation of CheA outdoes the negative feedback effect of CheB. If stimuli decrease, then the effect of CheB starts winning out, too few CheY proteins are phosphorylated, and the flagellum rotation reverts to its default rotation.
This control system alters the period between tumbles, with longer periods occurring in the presence of increasing attractants to prevent unneeded redirection.
Proteins as Gears
Schulz also explores the mechanisms by which the flagellum generates torque to spin the filament. While this process is not fully understood, he notes that the power output of the motor “is nearly 100% efficient,” and proposes that it involves proteins that effectively function as gears:
MotA in effect forms a “gear” and always rotates in the same CW direction. During normal CCW rotation for forward motion, the C-ring’s FliG engages with the side of MotA that is nearer to the C-ring axis. During CW rotation for tumbling, the MotA reconfigures so the C-ring engages with the side of the MotA cog gear that is farther from the C-ring axis. If this hypothesis is correct, the protein configurations of FliG and of MotA are exquisitely matched.
Schulz then turns to the assembly of the flagellum and notes that they have remarkably similar control across different species: “All flagellar systems coordinate flagellar gene expression through a transcriptional hierarchy central to an integrated regulatory network of multiple regulatory components. These networks exhibit a number of conserved circuit architectures reflective of the strong conservation found within the structural components of the flagellum.” In a series of illustrations he diagrams the flagellar assembly process through various pathways and at major stages in the production of various subcomponents. He notes that the complexity of the flagellum poses a challenge to evolutionary biologists:
The future work for an evolutionary biologist is twofold: (1) to provide a detailed explanation for how all the tightly constrained interlocking coherence described above could have evolved stepwise and naturalistically under real-world constraints; (2) to show evidence that such a scenario actually occurred in the past.
A Step-by-Step Pathway?
After concluding his review, Schulz expresses the view that a step-by-step evolutionary pathway is unlikely:
[T]he evolutionary biological community has yet to hypothesize a likely, detailed, step-by-step scenario to explain how the flagellum and its control system could have been blindly engineered naturalistically. Yet even that would still fall short of real evidence that such a thing actually happened, given real-world constraints. The flagellum seemingly is irreducible. How would portions of an incomplete, nascent flagellum be protected from degradation for generations while the remainder was yet to be gradually added? If some of the subassemblies discussed above could be omitted, what function would result?
Although Schulz notes that these are “hard” questions, he says they should not be dismissed: “These are real questions, and the challenge is to answer them.” In the meantime, despite our lack of total knowledge, he concludes that a design-option should remain on the table: “it seems disingenuous to dismiss teleology and intelligent causation, when so much is already known about the apparently ingenious, coordinated hierarchical assembly, control, and function of the flagellum.”
