 Evolution
Evolution
 Intelligent Design
Intelligent Design
If Malaria Is Evolving, It’s Not Doing It Darwin’s Way
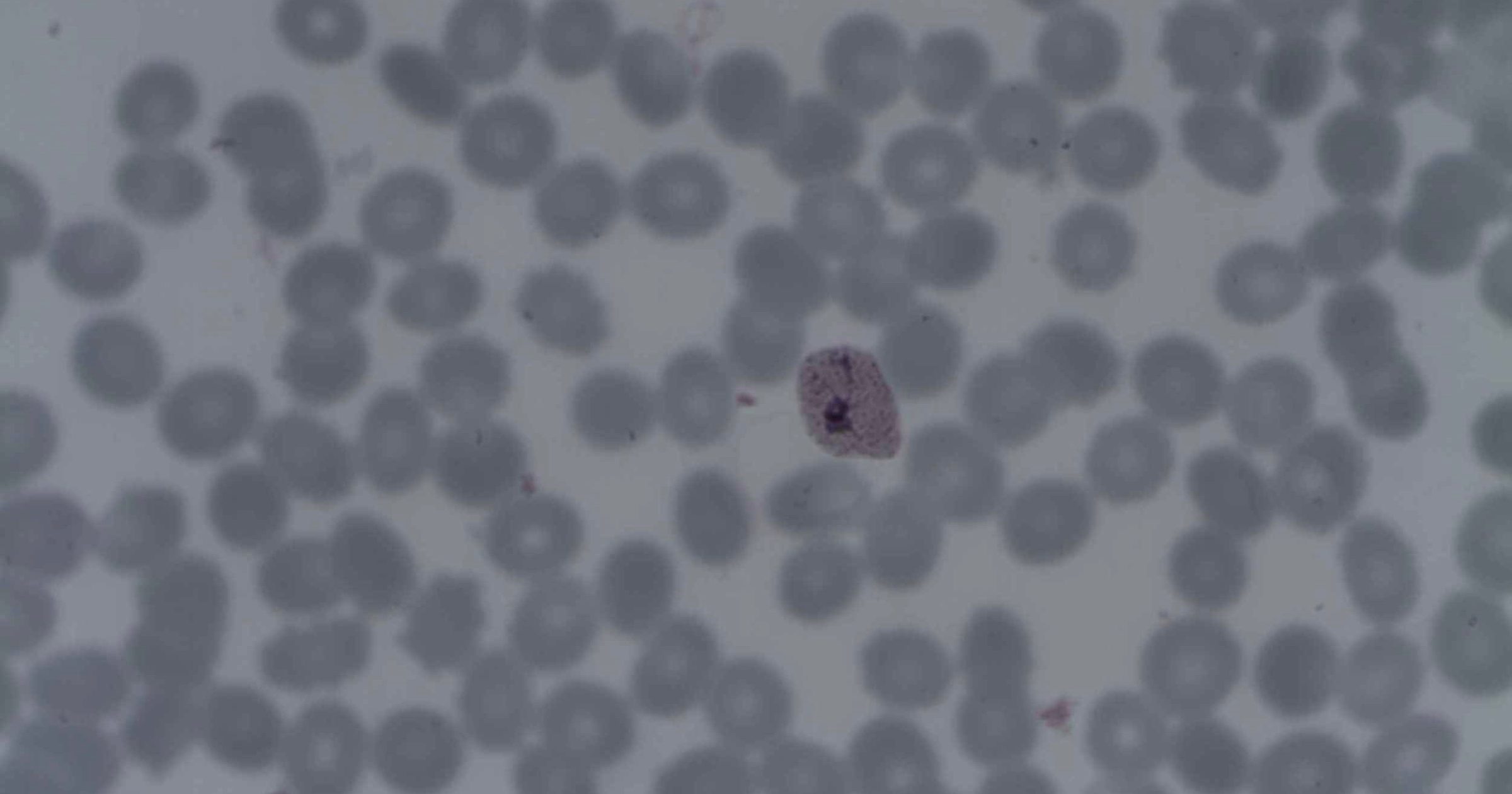
Here’s a chance to test Michael Behe’s book The Edge of Evolution. In the book, he showed the difficulty of waiting for coordinated mutations in the malaria parasite Plasmodium. With its billions of microbes living within billions of hosts, malaria offers abundant opportunities to test neo-Darwinism. The emergence of resistance to a drug that requires one mutation can be expected to occur regularly. Resistance that requires two coordinated mutations takes much longer. Three coordinated mutations, however, are so improbable as to never occur.
Since chloroquine resistance requires two mutations, it does occur from time to time. Beyond that, a drug will be past the edge of evolution. Like a soldier reaching safety beyond a mine field, a drug requiring three or more coordinated mutations will likely be immune from the emergence of resistance.
Evolutionary biologists at the Texas Biomedical Research Institute think they are “Catching malaria evolution in the act.” Using new single-cell sequencing techniques, they were able to monitor the arrival of new mutations in P. vivax. It looks like a challenge to Behe’s theoretical edge of evolution:
Researchers can now detect brand new mutations in individual malaria parasites infecting humans. Such high resolution could help us understand how parasites develop drug resistance and evade immune responses, and suggest potential treatment targets. [Emphasis added.]
They claim to have watched the evolution of drug resistance within two species of malaria, “Plasmodium falciparum, which is the deadliest; and Plasmodium vivax, which is the leading cause of recurring malaria infections because it can lie dormant in the liver and reemerge later.”
Using a high-powered magnet, they were able to isolate infected cells, which turn slightly magnetic when P. vivax invades young red blood cells. A laser beam then verified if the cells were infected. Running the infected cells through a flow cytometer, they could then use single-cell sequencing to look for new mutations. What they found did not fit neo-Darwinian predictions. Ian Cheeseman, co-leader of the study, explains:
“We would expect these brand-new mutations to be scattered randomly throughout the genome,” Cheeseman says. “Instead, we find they are often targeting a gene family that controls transcription in malaria.”
Already the neo-Darwinian picture is looking suspect.
But that’s not the only notable thing about the results. What really excites Cheeseman is that when the team compared single cell sequencing data for P. vivax and P. falciparum, the same transcription gene family contained the majority of new mutations for both species.
Behe sharpens his pencil as these findings are announced, because he feels a rebuttal coming for the claim of “catching malaria evolution in the act.” Mutations are supposed to be random. They should occur anywhere in the genome. Why are the majority of new mutations appearing in a gene family that controls transcription? Why are they appearing in two species not in contact with each other?
Astute readers should take note that sounding excited is a coping mechanism for evolutionary biologists when findings don’t match expectations.
Whatever is going on, it doesn’t look like random variation that Darwin expected to be the feedstock of natural selection. And the mention of plural mutations looks like what is happening is beyond the edge of evolution. Surprised by the findings, these neo-Darwinists pull out their tried-and-true excuse:
“We have two different species of malaria from two different parts of the world, Thailand and Malawi,” he says. “When we see the same thing happening independently in different species, this is an example of convergent evolution.”
When in trouble, when in doubt, run in circles, scream and shout. If coordinated mutations in one species are improbable, how does it help to posit similar coordinated mutations in two species?
Lost in Darwin Space
The researchers admit ignorance of what these mutations are doing. Their work has taken an unexpected turn.
The team does not know yet what impact the mutations have on the parasite and its ability to persist and cause damage in human hosts. The mutations may be critical for survival, or something like drug resistance, or may reveal those genes are unimportant.
“We don’t know what these mutations are doing,” Cheeseman says. “But the fact that they are targeting what is seen to be a fairly fundamental part of the parasite lifecycle is interesting and worthy of a lot of follow up.”
These preliminary findings are published in Cell Host & Microbe, “Single-genome sequencing reveals within-host evolution of human malaria parasites.” The Abstract summarizes the methods used:
We used single-cell sequencing protocol for low-parasitaemia infections to generate 406 near-complete single Plasmodium vivax genomes from 11 patients sampled during sequential febrile episodes. Parasite genomes contain hundreds of de novo mutations, showing strong signatures of selection, which are enriched in the ApiAP2 family of transcription factors, known targets of adaptation. Comparing 315 P. falciparum single-cell genomes from 15 patients with our P. vivax data, we find broad complementary patterns of de novo mutation at the gene and pathway level, revealing the importance of within-host evolution during malaria infections.
Clearly, understanding the observations is going to require “a lot of follow up” — but what kind?
Enter ID Scientists
Design-aware biologists with some expertise in engineering have an opportunity here. Now that the mutations appear non-random and clustered around similar genes controlling transcription, a working hypothesis could be that the variants are indeed providing a benefit to the organism. Here are some follow-up questions:
- Is there a sensor that triggers a mutagenic process when the cell is exposed to a new threat, like a drug it has not encountered before or an immune system that tries to fight it?
- Does the mutagenic process target genes that are able to respond to unforeseen threats? If so, what do those genes do?
- Is the mutagenic process a search algorithm for variations that provide protection? If so, is there a monitoring process that selects the variant and amplifies it in the host?
- How many coordinated mutations are found in a successful variant? (This can inform drug discovery able to surpass the edge of evolution.)
If indeed the findings point to a non-random process at work, this would indicate foresight, a hallmark of a designing mind. Designing a cell that can recognize and respond to unforeseen threats required good engineering. Whether malaria was designed to cause harm originally or devolved into what it is now is more a philosophical or theological issue, but findings from design-based research in this situation could help all microbiologists. It could begin to nudge them away from the default appeals to neo-Darwinism and convergence, and start them thinking about how an engineer would design cells for robustness in a dynamic world.
Editor’s note: See also Mike Behe’s recent article here, “Devolution Watch: Malaria Gnaws Off a Leg.”
