 Evolution
Evolution
 Intelligent Design
Intelligent Design
DNA Packing: One of the Supreme Wonders of Nature
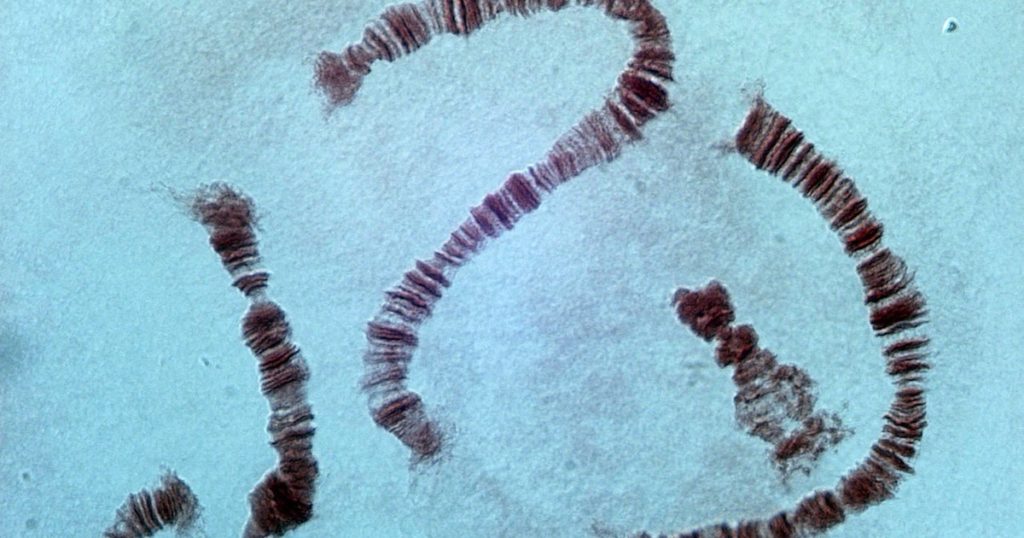
Chromosomes are densely packed DNA. The two “sister chromatids” of a chromosome, having been accurately duplicated during prophase and secured by centromeres, are arranged with all the other chromosomes on the spindle axis in metaphase. Soon after they are winched apart in anaphase into daughter cells. This elaborate choreography takes place every time a cell divides. The cell cycle is fascinating to anyone who has witnessed it under a light microscope, as you can see here:
But how did the chromosomes form before the cell cycle begins? Therein lies a true story of mind-boggling specified complexity that biochemists are still trying to understand.
Hand-Over-Hand Wrapping
Most people have had to wrap electrical cords or rope from a tangled mess into an orderly folded state for storage, using hand-over-hand motions. Scientists have known that in the nucleus, a molecule called cohesin is involved in wrapping DNA into chromatin (the building blocks of chromosomes). The loops formed by cohesin will be coiled and supercoiled further before maturation. The traditional picture of cohesin was that it traversed DNA strands like a sliding ring encircling the strand. That picture has been revised, reports Ruth Williams for The Scientist. It operates more like the human way of wrapping cords hand-over-hand:
Approximately 2 meters of DNA is crammed into each of the human body’s cell nuclei, themselves only about 10 μm in diameter. To achieve this packaging feat, DNA is wrapped around proteins to make chromatin fibers, and further condensed into tightly concertinaed loops. The formation and maintenance of these loops largely depends on two protein complexes — cohesin and condensin — each of which takes charge at different phases of the cell cycle. But how these complexes wrangle the stringy chromatin into submission has to this point been a bit of a black box.
Cohesin and condensin are ring-shaped like donuts, leading researchers to propose that chromatin might somehow thread through their middles. But work by chromatin biologist Jan-Michael Peters of the Research Institute of Molecular Pathology in Vienna, and colleagues now shows that, in cohesin’s case at least, the protein complex grabs DNA and pulls it, passing it from one part of itself to another, much like a person hauling rope might pass it hand to hand. [Emphasis added.]
How does a molecule pass a strand hand to hand without hands or eyes? The Scientist posted an infographic to show what Peters and team found. Cohesin has a “hinge” domain that fastens onto the DNA. The hinge moves toward one of two “head” domains. The other head clamps onto the strand several base pairs down, freeing up the hinge to reset and move down to another position.
There must certainly be more going on, because this doesn’t explain what happens to the upstream section that was reeled in. The open-access paper in Cell by Bauer et al., “Cohesin mediates DNA loop extrusion by a ‘swing and clamp’ mechanism,” says that each swing of the hinge forms a loop of DNA (see last April’s article on loop extrusion). Multiple loops parallel to each other begin the process of arranging the DNA for dense packing. Another player called NIPBL associates with cohesin into a cohesin-NIPBL complex during each 50-nm step, spending an ATP on each swing.
Here, we have analyzed how loop extrusion is mediated by human cohesin-NIPBL complexes, which enable chromatin folding in interphase cells. We have identified DNA binding sites and large-scale conformational changes that are required for loop extrusion and have determined how these are coordinated. Our results suggest that DNA is translocated by a spontaneous 50 nm-swing of cohesin’s hinge, which hands DNA over to the ATPase head of SMC3, where upon binding of ATP, DNA is clamped by NIPBL. During this process, NIPBL “jumps ship” from the hinge toward the SMC3 head and might thereby couple the spontaneous hinge swing to ATP-dependent DNA clamping. These results reveal mechanistic principles of how cohesin-NIPBL and possibly other SMC complexes mediate loop extrusion.
Remarkable movies made with super-resolution atomic force microscopy show the parts of cohesin undergoing conformational changes. These hand-over-hand motions operate in the dark without eyes, using ATP for energy. They get it right every time!
Fractal Organization
Our familiarity with loops of rope or cords may not translate to the structure of a completed chromosome. At the nanometer scale, one must take into account the attraction of molecules, including DNA, proteins, water, and other parts of the mix. The loops and supercoils of chromatin participate in forces of attraction and repulsion, confusing the neat hierarchical pictures of chromosome packing shown in textbooks.
Researchers at Postech Physics in Korea used a synchrotron, X-rays, and cryo-electron microscopy of intact chromosomes to peer deeper into the resulting package. Their results suggested a different model of organization: a fractal model.
The packing mechanism that condenses the chromosomes into one-millionth its size without any tanglingand the 3D structure that enables this have puzzled researchers for over a half a century. However, it has been difficult to observe the chromosomes in their native condition. The researchers had to resort to detecting only some components of the chromosomes or infer their condensed state from looking at their uncoiled state….
Through the study, the research team confirmed that the chromosomes were formed in a fractal structure rather than the hierarchical structure stated in previous studies. In addition, a physical model showing the packing process of chromosomes was presented.
Details of this work were published in PNAS by Sung et al., “Stochastic chromatin packing of 3D mitotic chromosomes revealed by coherent X-rays.”
Movies in the paper show that the molecules are constantly moving and vibrating, as would be expected with atoms and molecules in close proximity. How the DNA strands stay together for the process of mitosis, involving the “puzzling compaction mechanism from a DNA molecule to a chromosome and its error-free unpacking into DNA molecules again” without becoming hopelessly scrambled in the process, must count as one of the supreme wonders of nature. Look how they describe the accuracy of DNA packing despite all the interactions and motions of the adjacent molecules:
DNA molecules are atomic-scale information storage molecules that promote reliable information transfer via fault-free repetitions of replications and transcriptions. Remarkable accuracy of compacting a few-meters-long DNA into a micrometer-scale object, and the reverse, makes the chromosome one of the most intriguing structures from both physical and biological viewpoints.
A Work in Progress
This brief look at some of the inner workings of DNA packing should serve to arouse our awe and fascination. Somehow, it all works! DNA gets duplicated, organized, separated into two cells, and life goes on. Questions about “how these 11-nanometer-scale molecular complexes are spatially organized to form micrometer-sized chromosomes” and back again are sure to inspire wonder and research for decades to come.
