 Evolution
Evolution
 Intelligent Design
Intelligent Design
Topoisomerase Origins Defy Darwinian Explanations
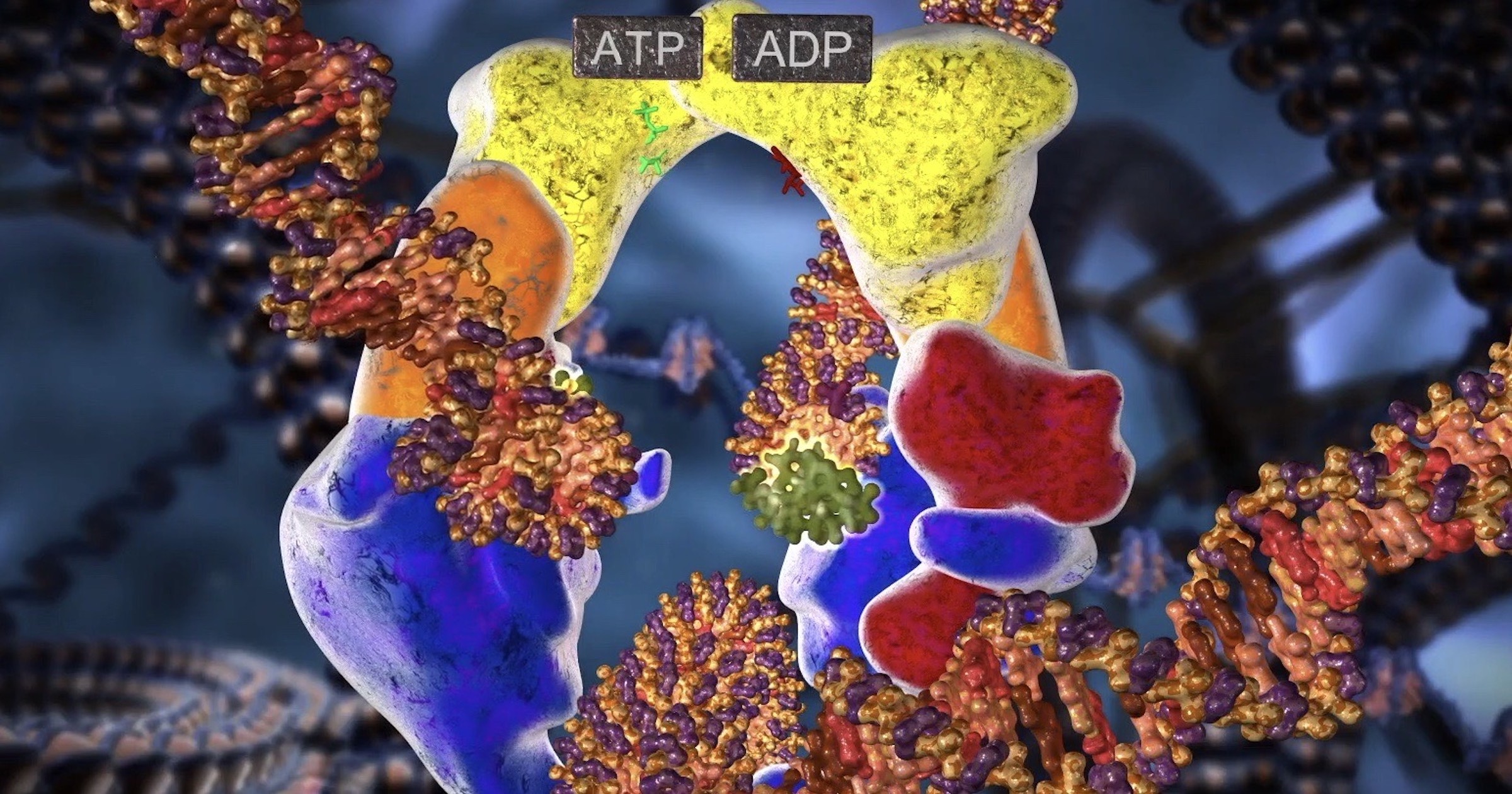
When Discovery Institute released the new animation of Topoisomerase II back in February, evolutionists could have sought to refute it. They could have submitted detailed accounts of how this multi-talented tool arose by a Darwinian process from simpler ancestors. Instead, they turned on their usual speculation machine.
A Review of Topoisomerases
Also in February, Yves Pommier from the National Cancer Institute (NCI) and three colleagues wrote a review article in Nature Reviews Molecular Biology about “Human topoisomerases and their roles in genome stability and organization.” This team no doubt wrote before the animation came out, and likely were unaware of it. Their review does not address origins; there is no mention of evolution, natural selection, or common ancestry. Neither is there any indication of intelligent design, although they mention molecular machines and motors six times.
First, let us marvel at the ubiquity and versatility of these molecular machines. It raises the question of whether any incipient life form could do without them, because even RNA molecules are subject to topological problems. An unstable genome is a dead genome.
DNA topoisomerases are present in all domains of life to resolve a wide variety of topological problems arising from the length of the human double-helix DNA polymer (about 3 × 109 bp) as it is folded, bent and highly compacted into the cell nucleus while remaining accessible to RNA and DNA polymerases. In addition, each human cell contains 100–1,000 copies of circular, ~16,000 bp mitochondrial DNA (mtDNA), as well as long and folded RNAs that are even more abundant than DNA and present in all subcellular compartments. [Emphasis added.]
There are six known topoisomerases in humans. “We discuss their specific and overlapping roles as regulators of nucleic acid topology and metabolism,” they begin, also indicating that failure in those roles often lead to cancer and other diseases (see Table 1 for a frightening list of syndromes caused by topoisomerase malfunctions: seizures, retardation, autism, premature aging, various cancers, and more).
To manage the topology of the long, folded and intertwined DNA and RNA polymers that are attached to scaffolding structures and are metabolically and dynamically processed by large molecular machines (such as transcription, DNA replication, chromatin remodelling and DNA repair complexes), human cells use their six topoisomerases often redundantly, but also in specific ways depending on the topological problem, the surrounding cellular structures and the differentiation status of the cell. This section outlines topological problems and the molecular solutions provided by each of the topoisomerases.
TOPS Are Always Needed
Topological problems are most likely to occur when the polymerases and helicases attach to DNA and RNA during key operations: transcription, replication, and chromatin remodeling among them. Operations that involve loop extrusion create risks for knots, over-twists, under-twists, catenanes, and other forms of torsional strain. These can lead to crossovers of the strands that can result in improper joining. Even single-stranded RNAs are subject to distortions in the genetic sequence. Operations that bring promoters and enhancers together, which can be considerable distances apart, can also lead to chromatin loops that require topoisomerase activity.
The six TOP machines are well equipped to handle specific emergencies, and they do not work in isolation; additional repair enzymes and cofactors must be at hand in all parts of the cell.
Human topoisomerases and the associated repair enzymes tyrosyl-DNA phosphodiesterases (TDPs; TDP1 and TDP2) are located both in the nucleus and in mitochondria; in addition, cytoplasmic RNAs are handled by TOP3B and TDP2.
Figures 1-3 in the paper show simplified diagrams of some common topological problems and illustrate which TOP machines work on them. The modes of action, though, as illustrated in the DI animation, are not discussed in detail in the review. The authors do speak of a “fine-tuned balance” between TopIIA and condensin during mitosis, pointing to another design requirement: just-in-time delivery of parts, reminiscent of assembly lines and operating rooms. Throughout replication and condensation of chromosomes during mitosis, the right TOP machines need to present at the right times and in the right quantities. Checkpoints ensure these requirements are met.
All mutations discussed in the review are disease-causing if not life threatening. Darwinians would wait in vain for a rare beneficial mutation, say from a UV ray, to “help” in any way. And if it occurred in a somatic cell instead of a germ cell, it would be but a brief and quickly forgotten stroke of luck.
Topoisomerases are magicians of DNA and RNA, and their full range of functions remain to be discovered.
How Can Evolutionists Darwinize All This?
Considering these facts, only a bold (or reckless?) individual would attempt to account for the origin of the topoisomerase system by unguided natural processes. One such attempt appeared in bioRxiv by Guglielmini and five co-authors, “Viral origin of eukaryotic type IIA DNA topoisomerases.” They start by recognizing the problem:
Type II DNA topoisomerases of the family A (Topo IIA) are present in all bacteria (DNA gyrase) and eukaryotes. In eukaryotes, they play a major role in transcription, DNA replication, chromosome segregation and modulation of chromosome architecture. The origin of eukaryotic Topo IIA remains mysterious since they are very divergent from their bacterial homologues and have no orthologues in Archaea.
And so, onward they proceed into the darkness of the mystery, looking for Darwin’s Cheshire-cat smile in the trees. But their entire approach is to look for homology between Type IIA topoisomerase genes and DNA sequences in certain viruses.
To elucidate the origin of the eukaryotic Topo IIA, we performed in-depth phylogenetic analyses combining viral and cellular Topo IIA homologs. Topo IIA encoded by bacteria and eukaryotes form two monophyletic groups nested within Topo IIA encoded by Caudoviricetes and Nucleocytoviricota, respectively…. The topology of our tree suggests that the eukaryotic Topo IIA was probably acquired from an ancestral member of the Nucleocytoviricota of the class Megaviricetes, before the emergence of the last eukaryotic common ancestor (LECA). This result further highlights a key role of these viruses in eukaryogenesis and suggests that early proto-eukaryotes used a Topo IIB instead of a Topo IIA for solving their DNA topological problems.
The homology argument, as shown in our video Long Story Short: Homology, is a circular fallacy: “Homology can’t be used as evidence for evolution because it assumes the very thing it’s trying to prove.” There’s also a personification fallacy in that last sentence. What in a proto-eukaryote, if such a thing existed, could decide it had a topological problem and would know how to latch onto a Topo IIB to solve it? And where did that handy tool come from?
With the root of the eukaryotic tree being still debated (Burki et al. 2019), it is difficult to propose a scenario for the evolution of Topo IIAs in eukaryotes. From our phylogenetic analyses, one cannot exclude that LECA already contained more than one Topo IIA.
Needless to say, their “scenario” presupposes the existence of Topo IIB and the viral genome to begin with. Some origin story! Begin with the thing already existing.
The eukaryotic molecular fabric appears to be a melting pot of proteins that originated in Nucleocytoviricota (mainly Megaviricetes), those that emerged de novo in the eukaryotic stem branch, proteins inherited from the bacterial ancestor of mitochondria and chloroplasts, and proteins that had ancestors in Archaea (in two domains scenarios) or in the common ancestor of Archaea and eukaryotes (in three domains scenario). Sorting out the viral component of our eukaryotic ancestors is now a major task in understanding eukaryogenesis.
This glimpse into the mental perspiration of Darwinians to account for the origin of exquisitely orchestrated factories of molecular machines serves a purpose other than entertainment. It shows that intelligent design remains the only serious contender for explaining the origin of life. We just need to get the word out. Sharing our animation on social media is a good way to participate.
