 Evolution
Evolution
 Intelligent Design
Intelligent Design
An Engineering Marvel: Uncovering the Mechanism of Respiratory Complex I
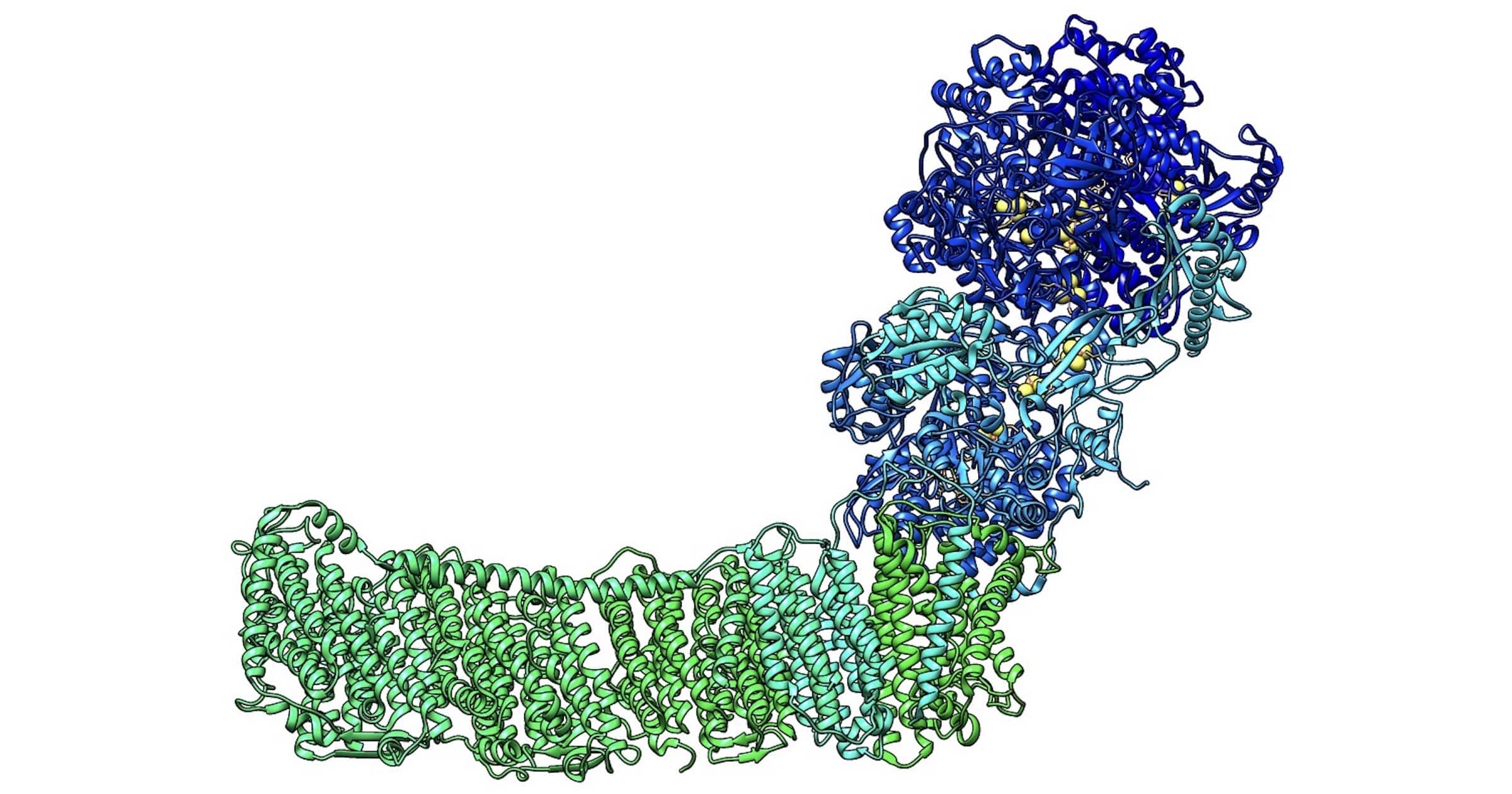
A couple of months ago, a friend recommended to me a 2011 paper on Respiratory Complex I, titled “Respiratory Complex I: ‘Steam Engine’ of the Cell?” The author is Leonid Sazanov, an eminent biochemist and structural biologist at the Institute of Science and Technology in Austria. This story quickly reeled me in. I binge-read, in about a week, the major papers detailing the unfolding mechanism of this crucial enzyme. The story has several plot twists including — spoiler alert — that Sazanov’s research group no longer thinks Complex I functions like a steam engine. Instead, they’ve discovered that enzyme electrical dynamics and conformational changes stack protons like dominos after which the Grotthuss mechanism or proton hopping occurs.
Complex I is involved in the electron transport chain, which is part of the biochemical process by which we create ATP, the energy molecule of life. Embedded in Complex I’s structure is advanced knowledge of the laws of physics, properties of electricity, and chemistry. Quite literally, Complex I is a nanoscale electrical converter that, in cooperation with other machines, charges one side of the membrane to create an electrical field that’s “equivalent to a bolt of lightning across every square nanometre of membrane.” (Somers 2022)
While my primary goal here is to describe the up-to-date understanding of the mechanism of Complex I, a close second goal is to kindle within my reader a sense of wonder about this enzyme’s amazing design.
The Basic Big Picture
Respiratory Complex I was a surprise to me, as it has been to its researchers. Sure, it is a proton pump, but how the protons are actually pumped… well that’s still up for debate — even after more than twenty years of intensive study.
Complex I is a key player in creating the mitochondrion’s electrical field. Complex I harnesses electron potential energy to push protons across the inner membrane to the intermembrane space. When positively charged protons pile up on the mitochondrion’s outer chamber, an electrical field of 30 million volts per meter is created. Yes, that membrane is crackling! In order to equalize this crazy charge separation, the protons rush back through molecular turbines or generators, called ATP synthase, spinning them in the process. This is what forms the body’s energy currency of ATP.
Complex I is present in nearly every living cell — from the lowly bacterium to the human mitochondria. Across species, Complex I shows similarities, but also differences. For example, mitochondrial Complex I has 30 more subunits than a bacterium’s Complex I — many of which are not understood. Sazanov, who has decades of experience studying the electron transport chain, thinks that all Complex I enzymes likely have the same basic mechanism. (Kravchuk et al. 2022) He seems to have identified what I will call a design motif. See Figure 1 below for a timeline of discoveries that lead to this conclusion:
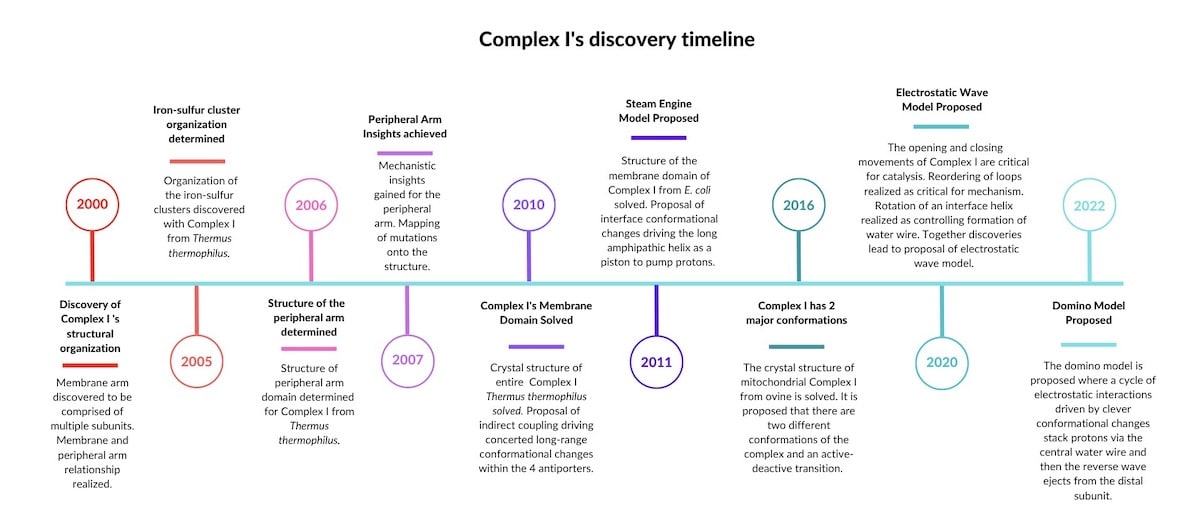
Sources for the timeline:
- 2000. “Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme.” PMID: 10852722
- 2005. “Organization of iron-sulfur clusters in respiratory complex I.” PMID: 16051796
- 2006. “Structure of the Hydrophilic Domain of Respiratory Complex I from Thermus thermophilus.” PMID: 16469879
- 2007. “Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain.” PMID: 17274631
- 2010. “The architecture of respiratory complex I.” PMID: 20505720
- 2011. “Respiratory complex I: ‘steam engine’ of the cell?” PMID: 21831629
- 2016. “Atomic structure of the entire mammalian mitochondrial complex I.” PMID: 27595392
- 2020. “The coupling mechanism of mammalian respiratory complex I.” PMID: 32972993.
- 2022. “A universal coupling mechanism of respiratory complex I.” PMID: 3610456
Introducing Complex I’s Mysteries
Complex I is a membrane protein. If you’ve done any protein crystallography, you will know that membrane proteins don’t like to have their structures determined. Not wanting to be an exception, Complex I resisted scientists’ attempts for years. It wasn’t until 2006 that structural breakthroughs started to happen.
Solving the structure of a protein usually reveals the mechanism of how the protein works. But with Complex I, this turned out not to be the case. Instead, structural characterization revealed a peripheral arm (PA) connected to a membrane domain (MD) forming something like an “L”-shaped structure (Figure 1, 2000). The peripheral arm was found to contain all of the redox centers (where electron flow happens), but the proton pumping channels (or what everyone thought were proton pumping channels) were too far away (a distance of over 200Å) for direct coupling. This puzzled everyone, since direct coupling between electron transfer and proton translocation had already been observed in other members of the electron transport chain or ETC (Complexes III and IV). While coupling can be indirect, this would involve conformational changes over a distance. In the case of Complex I, neither were observed. There were no large-scale conformational changes obvious from the structure and the redox centers were far (200Å) from the location of proton translocation. You might say that everyone was scratching their heads.
Emerging Knowledge about Complex I’s Redox Centers
Fairly early on, it was known that to start Complex I’s cycle, which culminates in pumping of four protons, a molecule of NADH (a mobile electron carrier) must deliver an electron pair (as a hydride) to the tippy top of the peripheral arm of Complex I.
There the electrons are transferred as a pair from NADH to FMN (flavin mononucleotide) — a molecule that typically wants electrons more than NADH. FMN is bound within Complex I’s peripheral arm and functions as a two-to-one electron converter, handing one electron to an upstream off-path N1a Fe-S cluster and one electron to the first Fe-S cluster (N3) in the path to ubiquinone.
Let’s Review the Basics of Electricity and Voltage
To fully appreciate redox reactions, and the next part of Complex I’s history, you’ll need to recall a thing or two about electricity. Electrical fundamentals start with the knowledge that electrons travel towards positive charge and that electrical potential is measured in volts (V). Voltage is just a measure of the pressure electrons find themselves in (how badly they want to go from point A to point B). Because of their desire to get from point A to point B, electrons can do work, or, specifically in Complex I’s case, attract protons.
Redox potential (the biochemist’s way of saying electrical potential) is measured in milliVolts (mV). If an electron is at a location with redox potential of -10 mV and there are two locations the electron could go towards, one with a redox potential of -8 mV and one with a redox potential of -12 mV, the electron will move toward the more positive location, i.e. -8 mV. Therefore, electron flow can be established by having a stepping stone path of increasingly positive potentials. This is what happens in Complex I. NADH hands electrons to FMN, which has a higher desire for electrons, and then FMN subsequently hands the electrons to the Fe-S clusters.
The Fe-S Cluster Pathway
There are eight Fe-S clusters in most versions of Complex I that the electrons step through (Figure 1, 2005). The potentials of the Fe-S clusters actually don’t have a stepping stone path of increasing potentials, but instead have a roller coaster redox potential profile (high to low potential) which is thought to optimize the rate of electron transfer and improve energy extraction efficiency. Some of these Fe-S centers are likely control centers. For example, one iron sulfur cluster acts like a bottleneck to control the overall rate of electron transfer (basically a resistor), while another is pH sensitive.(Sazanov 2015) The only significant redox potential drop occurs in the final transfer of the electrons to the acceptor molecule quinone. This is a hint that the crucial energy releasing step (~100 mV) is quinone reduction or even the release of quinone out of the cavity.
Quinone Acts as an Electron Acceptor
Quinone (also called “CoQ10”) can act both as a single electron carrier and a double electron carrier, making it the perfect 1-to-2 converter for the Fe-S clusters. It has a quinone head group and ten isoprene tail groups (in mammals). This makes it very hydrophobic (think: avoids water), and sure enough it spends its life in the hydrophobic tails of membrane lipids. In Complex I there is a shallow binding site and a deep site for quinone. The opening for quinone to enter is within the membrane. However, on the opposite side from where quinone enters there is a hole in the peripheral arm called “W” where water is free to enter the quinone binding cavity from the matrix or cytosolic side. The binding cavity for quinone is also lined with hydrophilic residues which repel a hydrophobic molecule like quinone — in this case pushing it into the deep binding site. Quinone’s entry and the “W” site close and open at the same time depending on the state of the enzyme’s catalytic cycle. When both are open the cavity is prefilled with matrix-derived H2O molecules, and quinone can enter the shallow binding site. As it moves deeper into the cavity it pushes the water out through the “W.” What’s happening here is the same thing that happens when you try to suck liquid out of a tightly closed container — at first, it’s difficult until you poke a second hole. The second hole, in this case “W,” allows the water to be easily displaced, making binding easier. As quinone moves deeper into the binding site, disordered loops become structured or ordered, closing the “W.” They also partially close the entry point sealing ubiquinone into the cavity. The conformational changes which take place connect the water wire running through the membrane domain to the peripheral arm, which we will discuss next.
The Maddening Membrane Domain
Until 2019 the membrane domain was described as having four proton pumps, but… plot twist… the newest research suggests that only the final subunit pumps. While the other subunits are necessary for a continuous water wire and pumping of four protons, they do not have outlets to the intermembrane or periplasm space.
Early Ideas About How the Membrane Domain Functions
- Idea #1: Homologous Antiporters?
In 2013, the long-awaited crystal structure of the membrane domain revealed antiporter-like subunits first said to be homologous to Na+/H+ antiporters and later specifically to the Mrp antiporters (Figure 1, 2013) (Efremov, Baradaran, and Sazanov 2010). A generic single subunit antiporter is a protein that swaps ions across a membrane. The homology-informed speculation to a generic antiporter was incorrect, in some sense, because now it has been shown that there is only one outlet by which protons leave the complex (not three or four). Furthermore, the Mrp complex is now thought to function like Complex I.
Why Design Triangulation Is Superior to Homology
At this point I want to make a little jab at homology (an evolutionary idea) versus design triangulation (an ID-based concept which predicts what parts might exist in a system in order to satisfy the necessities of expected design specifications). When comparing two proteins in terms of structure or sequence, if similar, evolutionary scientists might say the proteins are homologous. The definition of homology is that two things exhibit similarity BECAUSE of their being evolutionarily related. However, often the reason an evolutionary scientist says this is NOT necessarily because they have a clear, undisputed path showing this relatedness, but because the structures have some similarity, and the scientist wants to infer that they do something similar.
However, this inference — that objects that appear similar in their coding or structure might have similar functions — does NOT require evolutionary theory. In fact, what is actually happening in “homology” comparisons is what philosopher of biology, Paul Nelson, calls design triangulation. When we see things that have a similar code or structure, we triangulate based on what we’ve seen before and on our knowledge of how it works. This does not require any inference about ancestry. We use design triangulation to infer that a newly discovered thing might do something similar to the previously discovered thing. Design triangulation is superior to the evolutionary-based homology inference because design triangulation also says something about the differences between the objects being compared. Design triangulation predicts that the differences between objects can give us a clue as to the uniqueness of the design for a particular function (something typically lost in the evolutionary homology-based comparison).
For Complex I, there were early clues about differences between generic antiporters and Complex I’s hypothetical homologous antiporters. (Sazanov 2015) Importantly, the researchers noted the presence of unexpected key lysine residues at the center of the half channel (the homology-informed speculation was anticipating carboxylates) and called them “highly unusual.” (Sazanov 2015) As it turns out, these key lysine residues would end up being important clues as to how charges are stacked in Mrp and Complex I’s mechanism. Thus, Complex I is a case in point where differences from homology-based speculations provided clues for a unique mechanism.
- Idea #2: The Transverse Helix and Steam Engine Idea
Another interesting feature revealed in 2010 by the long-awaited crystal structure was the presence of a long amphipathic helix running through the membrane domain. (Efremov, Baradaran, and Sazanov 2010; Efremov and Sazanov 2011b) This helix was initially proposed to act like a piston to pump protons, hence the steam engine paper. (Efremov and Sazanov 2011a) But further structural characterization of cow and sheep versions of respiratory Complex I did not reveal major conformational changes necessary for this type of mechanism.
- Idea #3: Electrostatic Waves?
A third idea was that electron transfer events facilitate conformational changes along the complex, manifesting like a forward and backward electrostatic wave.
Discovery of the Transverse Water Wire
Instead of any of these hypotheses being confirmed, what began to come into focus as structural resolution improved is that a water wire ran transversely along rather than across the membrane domain, with two openings on the matrix side and one opening to the intermembrane space. The water wire was connected and disconnected to the redox center by a rotating helix as the enzyme opened and closed (during turn over or catalytic cycle).
Water wires are single file arrangements where protons can be ferried from one water molecule to the next extremely rapidly. Another name for this phenomenon is a Grotthuss chain. Here’s how it works: When a proton approaches a water wire, there is cooperative breaking of hydrogen bonds and reforming of a covalent bond until the end of the wire is reached. At that point the last water molecule of the chain ejects a proton out, completing the proton conduction.
Scientists Figure Out How the Enzyme Works: A Domino Model
With the discovery of the water wire, and the knowledge that Complex I cycles through open and closed states, the scientists put together a model for how the enzyme functions. They call their model the domino model. In short, the “domino effect” refers to a series of Grotthuss chain proton transfers. Quinone reduction causes a forward wave of “domino stacking” whereby protons are pulled into the central axis from the cytosol, or cytoplasmic matrix. Then, electrostatic interactions (protons repulsing each other) initiate a proton vacancy at the other end of the Grotthuss chain which initiates the reverse wave like “dominoes falling,” resulting in pumping of four protons into the intermembrane space. A cycle of Complex I results in the pumping of four protons and can be broken down into five stages. See Figure 2 below which is adapted from Kravchuk et al. 2022.
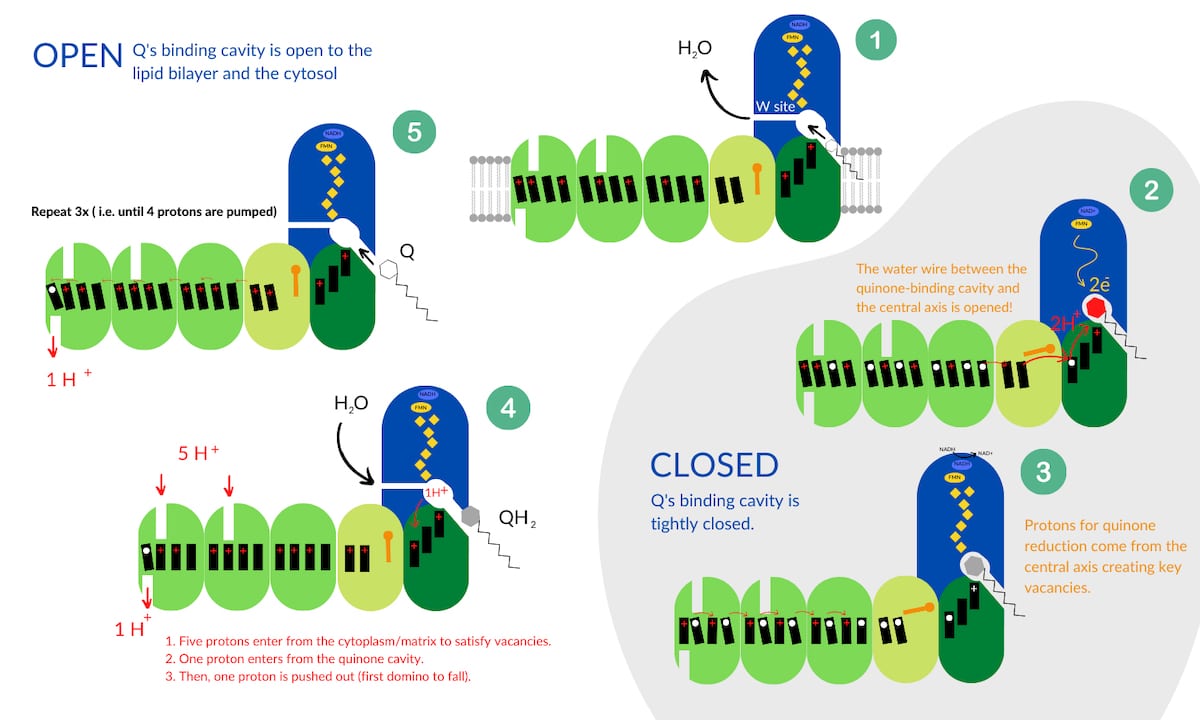
- Stage 1
Complex I starts its cycle in an open state. The open conformation means that quinone’s binding cavity is open to the lipid bilayer and the cytosol (Stage 1 in Figure 2). The “W” site is open, allowing water to move out of the cavity as quinone moves in. Initially the charges in the central axis are not stacked (depicted as tilted black rectangles).
- Stage 2
When quinone binds, the complex moves to a closed state and quinone gains the two electrons arriving from the Fe-S clusters. To finish this reaction, two protons are required. At this point quinone is trapped and closed off from exterior proton sources including the “W” site or where it entered. However, when quinone moved up into the binding site, conformational changes connected the transverse water wire between Complex I’s central axis and quinone’s binding cavity (depicted by an open orange gate in Figure 2). Thus, protons from the central axis flow up into the binding cavity to complete quinone’s reduction. However, this creates vacancies in the central axis (white circles).
- Stage 3
As the water wire delivers protons to satisfy the negative charges on quinone, protons from the central axis flow to the right (Stage 3 in Figure 2) to satisfy the vacancies (white circles). This of course creates more vacancies at key positions in the central axis. This movement is considered “domino stacking” (note the upright black rectangles at charge destinations).
- Stage 4
As the enzyme transitions to the open state, five protons from the cytosol and one proton from the cavity rush in to protonate the vacancies created in Stage 3. Reduced quinol is kicked out of the cavity, and conformational changes disconnect the water wire, leaving charge tension in the membrane domain. To relieve the tension, one proton (aka, the first domino to fall) is ejected into the periplasm, or intermembrane space, and creates a vacancy.
- Stage 5
That vacancy sets off a cascade of proton replacements in the central axis. This results in another vacancy, and protons start being redistributed to fill the vacancy. Eventually all the tension is relieved by pumping four protons into the intermembrane space. The ultimate result is that a powerful electrical potential accumulates on one side of the membrane which can be used to spin ATP synthase.
An Inference to Intelligent Design
Now you see why it is not an overstatement to call Complex I an engineering marvel. This 1.6605 e-18 gram machine’s mechanism has been worked out piece by piece over more than twenty years, revealing cooperation between nanoscale chemistry, physics, and electrostatics. While the best engineers today can’t build something like this, and it took the brightest minds a couple of decades to discover its tightly held secrets, evolutionary scientists still attribute this remarkable design to random mutation and natural selection. Whatever your views on how life and Complex I came to be, I hope this description provides perspective on why other scientists, myself included, believe that life was intelligently designed.
From the strategic placement of multiple Fe-S clusters to the design of a specific quinone binding pocket with a resealable hole, when I look at Complex I, I see remarkable design. Thirteen separate genes encode the minimal bacterial Complex I, all of which must interface specifically so that when assembled, highly efficient proton transfer can occur. The hypothesis that Complex I came about by co-option of other biochemical parts, when it must have been so central so early in the history of life, is a bit preposterous. Consider also how the function of this enzyme relies upon foresight of the cellular context, i.e., a membrane, FMN, and CoQ10. Can natural selection move multiple pieces in parallel towards a unified goal? I have not seen a plausible explanation of how it can. Is it reasonable to expect gradual evolution to be responsible for the specified residues that enable a Grotthuss mechanism such that when one proton moves it bumps the next?
I think we may safely infer that Complex I was designed, and that is for the following reason. When we see a power adapter allowing electron transfer between your laptop and the wall outlet, we attribute it to design. When we look at the Hoover Dam as it utilizes potential energy to generate electricity, we attribute that to design. Yet, Complex I’s engineering marvels surpass both a power adapter and the Hoover Dam’s generators. Because this and many other molecular machines exhibit features that, in the context of human technology, we naturally attribute to design rather than to happenstance, we should be consistent and also infer that molecular machines are the results of intelligent design.
Sources:
- Efremov, Rouslan G., Rozbeh Baradaran, and Leonid A. Sazanov. 2010. “The Architecture of Respiratory Complex I.” Nature 465 (7297): 441–45.
- Efremov, Rouslan G., and Leonid A. Sazanov. 2011a. “Respiratory Complex I: ‘Steam Engine’ of the Cell?” Current Opinion in Structural Biology 21 (4): 532–40.
- Efremov, Rouslan G., and Leonid A. Sazanov. 2011b. “Structure of the Membrane Domain of Respiratory Complex I.” Nature 476 (7361): 414–20.
- Kravchuk, Vladyslav, Olga Petrova, Domen Kampjut, Anna Wojciechowska-Bason, Zara Breese, and Leonid Sazanov. 2022. “A Universal Coupling Mechanism of Respiratory Complex I.” Nature 609 (7928): 808–14.
- Sazanov, Leonid A. 2015. “A Giant Molecular Proton Pump: Structure and Mechanism of Respiratory Complex I.” Nature Reviews. Molecular Cell Biology 16 (6): 375–88.
- Somers, James. 2022. “How Food Powers Your Body.” The New Yorker, October 25, 2022. https://www.newyorker.com/science/elements/how-food-powers-your-body-metabolism-calories.
