 Intelligent Design
Intelligent Design
The Cell’s Surveillance System: Introducing the Cell Cycle Checkpoint Pathways

In previous articles, I’ve reviewed some aspects of the incredible design of the eukaryotic cell cycle (see here and here). 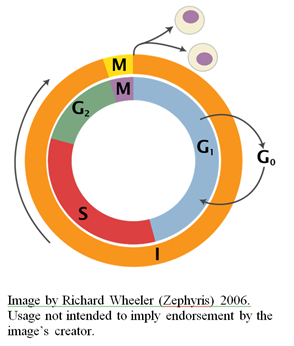 I would like to turn now to considering the various cell-cycle checkpoints, so-named by Leland Hartwell and Ted Weinert in 1989 (Hartwell and Weinert, 1989).
I would like to turn now to considering the various cell-cycle checkpoints, so-named by Leland Hartwell and Ted Weinert in 1989 (Hartwell and Weinert, 1989).
The checkpoint pathways can be thought of as the cell’s surveillance systems that function to arrest cell cycle progression in response to detected problems in cell division or chromosome replication. Checkpoint pathways ensure that DNA replication does not commence before all of the components necessary for its completion have been produced. They also ensure that the replication process is not derailed by damaged DNA; that cells do not attempt to divide before genome duplication is complete; and that each daughter cell receives a complete set of chromosomes. Checkpoint pathways are essential for an organism’s viability. Although a cell can complete its cycle successfully without them, their absence results in infidelity of chromosome transmission and a greatly raised susceptibility to DNA-damaging agents. The consequence of checkpoint pathway inactivation is genome instability, which inevitably leads to cancer (e.g. Chiu et al., 2012).
Introducing the Checkpoint Pathways
The video above provides a basic introduction to the various checkpoint pathways that monitor the cell cycle as it progresses from one phase to the next. The activation of checkpoint pathways involves proteins called sensors, transducers and effectors. Sensors detect the cellular defect and trigger an appropriate response. Sensors are specific to the type of damage, and this ensures that the correct response is initiated. DNA damage, for instance, is typically characterized by the formation of single-stranded DNA (ssDNA), in which case the activation of the corresponding checkpoint pathway is dependent upon the binding of sensors to ssDNA (Zou and Elledge, 2003).
Sensor proteins activate transducer proteins. Transducers function to initiate the appropriate response to damage. Transducers include protein kinases and proteases, and serve to activate the effector proteins (which carry out the response itself). Transducers that are activated in response to damaged DNA, for example, will in turn activate effector proteins that play a role in DNA damage repair, production of nucleotides and cell cycle inhibition.
The Restriction Checkpoint Pathway
As I mentioned in a previous article, commitment to another round of cell division depends on a number of conditions, such as the presence of sufficient nutrients to support an increase in mass and synthesis of the proteins and nucleotide precursors necessary for genome duplication. The point at which a mammalian cell irreversibly commits itself to another round of division is known as the “restriction point,” a term first coined in 1974 by Arthur Pardee (Pardee, 1974). The equivalent in yeast cells is called the “start point.” Without the necessary conditions for passing this point, eukaryotic cells enter into the so-called G0 state where cell proliferation does not occur. Whether a cell commits to another round of proliferation or not is determined by the restriction checkpoint pathway.
The key Cdk inhibitor at the restriction point is p16, which acts to inhibit Cdk4/6 and prevents them from binding to cyclin D to facilitate cell cycle progression (Villaca�as et al., 2002; Russo et al., 1998; Serrano et al., 1993). When growth is induced, however, the increased expression of cyclin D allows it to overcome p16 and competitively bind to Cdk4/6. Upon formation of Cdk4/6-cyclin D complexes, the tumor suppressor Rb (which is inhibitively bound to the transcription factor E2F) is phosphorylated, thus freeing E2F which subsequently triggers cyclin E expression (Weinberg, 1995; Matsushime et al., 1994; Meyerson and Harlow, 1994; Kato et al., 1993). When cyclin E binds to Cdk2, it triggers the transition from G1 to S phase.
For a recently published review of this checkpoint pathway, see Bertoli et al. (2013).
The DNA Damage Checkpoint Pathway
During replication, the DNA duplex is separated into two strands, each of which serves as a template for the synthesis of a novel strand (referred to as “semi-conservative” replication). Sometimes, however, replication can be impeded by an inadequate supply of nucleotides, or a double-strand break (resulting in a stalled replication fork). In such cases, DNA replication comes to a halt, leading to the accumulation of single-stranded DNA. Single-stranded DNA is bound by sensor proteins, which trigger the activation of the checkpoint pathway (Zou and Elledge, 2003). The sensor proteins activate transducers (protein kinases), which activate various effectors. This in turn leads to cell cycle arrest and DNA repair (Jackson, 2002).
In the case of partially replicated DNA, in response to detected DNA damage, progression of the cell cycle will halt in S phase, and mitotic cyclin-Cdks will be inhibited to ensure that mitosis does not begin with only partially replicated DNA (the consequences of which would be catastrophic — with the daughter cells inheriting incomplete genomes). When DNA damage is detected in the S or G2 phase, mitosis is blocked by preventing the activation of mitotic cyclin-Cdk, e.g. by down-regulation of mitosis-promoter Cdc25 or up-regulation of mitotic inhibitor Wee1 (Raleigh and O’Connell, 2000). The DNA damage checkpoint pathway can also arrest cell cycle progression if DNA damage is detected in G1 phase, and inhibit the activity of G1-cyclin-Cdk (thereby blocking passage through the restriction point).
Checkpoint pathways can also on occasion bring about programmed cell death (apoptosis) in cases where damage cannot be properly repaired (Gartner et al., 2000). See Musaccio and Salmon (2007) for a review of the DNA damage checkpoint pathway.
The Spindle-Assembly Checkpoint Pathway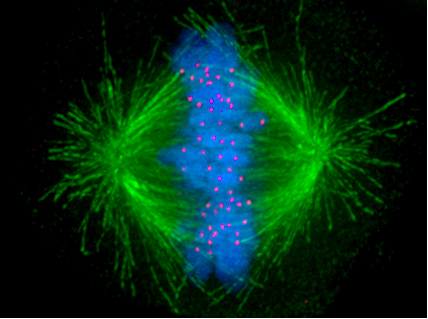
Eukaryotic cells also have a checkpoint dedicated to monitoring chromosome segregation during mitosis (M phase). In a previous article, I introduced the mitotic spindle which attaches to chromosomes at structures called kinetochores and pulls them to the poles of the cell where they segregate into daughter cells. This in itself is a truly astonishing process — one that I intend to discuss in further detail in a future article.
Of critical importance to chromosome segregation is the correct attachment of the spindle fibers to the chromosomes. The consequences of improper attachment can be devastating, with both copies of a chromosome being segregated to only one daughter cell. The role of the spindle attachment checkpoint pathway is to monitor the attachments between the spindle fibers and chromosomes (Lara-Gonzalez et al., 2012).
The spindle-assembly checkpoint is active during M phase of the eukaryotic cell cycle, and prevents dividing cells from progressing beyond metaphase by inhibition of the anaphase-promoting complex (APC) until the chromosomes have been properly attached to the spindle fibers (Rossio et al., 2013; Foster and Morgan, 2012; Lara-Gonzalez et al., 2012; Nilsson et al., 2008). Establishing of proper attachment results in the extinguishing of the checkpoint signal, and the cell continues to progress through mitosis. There is evidence to indicate that signaling in response to unattached or improperly attached kinetochores, in addition to tension sensing, contributes to this checkpoint pathway (e.g. see Uchida et al., 2009; Zhou et al., 2002).
Conclusion
My discussion thus far of the molecular mechanisms underpinning cell cycle progression has barely scratched the surface, and indeed entire books have been written on the subject (such as this one). The more deeply you explore the cell cycle, however, the more you come to the realization that it represents one of the most remarkable evidences of design in the biological universe.
