 Intelligent Design
Intelligent Design
 Life Sciences
Life Sciences
Foresight in Single Cells
In his book Foresight: How the Chemistry of Life Reveals Planning and Purpose, the wonderful Brazilian chemist Marcos Eberlin gives many examples of phenomena that only make sense if a designing intelligence saw a need and planned for it in advance. In Chapters 3 and 4, he introduces some of the features of genes and enzymes as examples of his thesis, stating on page 141:
The need to anticipate — to look into the future, to predict potentially fatal problems with the plan, and solve them ahead of time — is observable all around us…. Put another way, many biological functions and systems required planning to work.
He could not begin to exhaust examples in a small book of 161 pages; it would require an encyclopedia — or a library. Here are some new findings that reinforce his idea of foresight within the microscopic world of the cell.
The Antenna Repair Garage

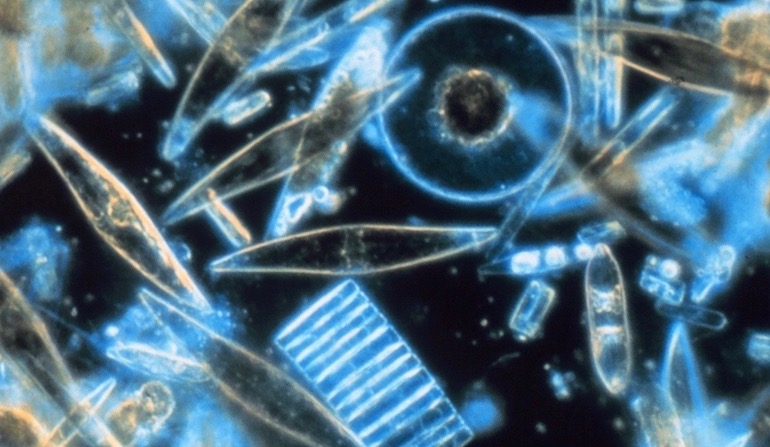
Diatoms are photosynthetic algae that live in glass houses of intricate geometric design and artistic beauty. Found worldwide in both salt water and fresh water, these autotrophs (organisms able to manufacture their own food) join plants and bacteria as generators of molecular oxygen for the rest of us. Inside diatom shells (called tests), chloroplasts contain the photosystems for this vital work. Within the thylakoid membranes of the chloroplasts, “Photosystem II” antennas capture sunlight and direct the energy toward ATP synthase rotary engines for ATP production, and toward other enzymes for carbohydrate production. While lacking the structural organization of grana and stroma seen in the chloroplasts of green plants, the diatom’s photosystems live in a highly energetic environment, where breakdowns can occur.
Looking inside the thylakoid membranes of a particular diatom, Orly Levitan and her team were intrigued to see two groupings of photosystem II (PSII) complexes in different locations. The first group was functioning properly, but the second group was silent, even though all the parts appeared to be present. They deduced that something amazing was going on. Writing in PNAS, they said:
Using cryo-electron tomography in conjunction with proteomic and biophysical analyses, we show the distribution of photosystem II in thylakoid membranes of a diatom is heterogeneous. There are 2 subpopulations of the PSII supercomplexes; one contains antennae complexes, and the other does not. The former is functional, while the latter appears to be photochemically inactive. We suggest, based on tomography and biophysical analysis, that photochemically damaged reaction centers are physically isolated to repair stations where the inactive proteins can be removed and replaced. [Emphasis added.]
The authors believe that repair locations have been specifically designated where “photochemically damaged proteins can be removed and replaced with functional structures.” Does that not look like a plan with foresight? The repair shops are physically isolated from the highly energetic environment of the working PSII complexes, just like pit stops that allow race cars to be dragged to safe places off the speedway for repairs.
As for the origin of this wonder, the authors only assert that “diatoms evolved.” Once again, the evolutionary miracle story requires additional miracles: “convergence” between diatoms and unrelated red algae:
The observed segregation suggests that despite the evolutionary differences between the red and the green eukaryotic algal lineages, there is convergence in the spatial segregation in distribution of the PSII supercomplexes. The spatial segregation implies a common functional process that was inherited before the evolutionary split of eukaryotic algae.
Note the contrast between “observed” and “suggests” in the quote, indicating that the evolutionary story is not derived from observations, but from ideology.
Skin Tight
Cells in their flimsy-looking membranes look like squishy blobs ready to fall apart at the slightest tug. Yet they are surprisingly robust. Red blood cells, for instance, travel through narrow capillaries like contortionists, then pop back into shape when the vessel widens. How is that possible? The answer is in an internal skeleton, the cytoskeleton, that is remarkably tough and flexible at the same time. Microbiologists have known about a couple of components of the cytoskeleton — actin and microtubules — but another class of players is emerging as critical.
An international team from MIT, Harvard, and other top universities found that “High stretchability, strength, and toughness of living cells [is] enabled by hyperelastic vimentin intermediate filaments” (PNAS).
Intermediate filaments (IFs) remain the least understood with respect to their functions in mammalian cells even though they have been related to many devastating human diseases. Here we use optical tweezers to perform micromechanical measurements in living cells and in IF enriched cytoskeletons devoid of actin and microtubules. We identify that cytoskeletal vimentin IFs (VIFs) provide cells with a hyperelastic rubber-like network that regulates the essential mechanical properties of mammalian cells including stretchability, strength, resilience, and toughness. We show that VIFs maintain cell integrity and viability under conditions involving extreme deformations. We further show that the stretchy VIF network can effectively disperse locally induced mechanical stress to larger regions within individual cells, enabling the dissipation of energy throughout a cell.
The team of Hu et al. determined these amazing properties by creating “ghost cells” of just the networks of intermediate filaments (IFs). This allowed them to measure the toughness and flexibility of the filaments as they tugged on them and put them through various contortions, including “large deformations.”
Our results show that cytoskeletal VIFs form a stretchable, hyperelastic network in living cells. This network works synergistically with other cytoplasmic components, substantially enhancing the strength, stretchability, resilience, and toughness of cells. Moreover, we find the hyperelastic VIF network, together with other quickly recoverable cytoskeletal components, forms a mechanically robust structure which can mechanically recover after damage.
Foresight in this instance can be seen by imagining the evolution of such a network in a stepwise manner. Suppose the VIF network was stretchy but not tough; it would break easily. Suppose it was tough but not flexible; it would not be able to “mechanically recover after damage.” Suppose the VIF network was unable to “work synergistically with other cytoplasmic components” – it would get in the way of other systems rather than enabling them. The entire network, with the four identifiable properties of the filaments (strength, stretchability, resilience, and toughness) all had to work synergistically with the entire cell from the beginning.
Thank these authors for not dragging readers through Darwin storybook land. Instead, they note with surprise that even after 100 cycles of deformations, “the cytoplasmic VIF network itself is hyperelastic and with negligible energy dissipation.” In fact, repeated deformation “causes most cytoskeletal structures to be damaged, disassembled, or rearranged in cells without VIFs.” All together, though, everything works perfectly.
As a highly stretchable hyperelastic network, the cytoskeletal VIF network greatly enhances several of the most important mechanical properties of the cytoplasm: stretchability, resilience, strength, and toughness (SI Appendix, Table S1). The cytoplasmic stretchability is mainly determined by the VIF network. Subjected to repeated loadings, the VIF network plays an essential role in maintaining the resilience of the cytoplasm because of its high yielding strain, while the rest of cytoplasmic components are greatly softened or even disassembled. By interacting with other cytoskeletal systems and organelles, VIF networks significantly enhance cytoplasmic mechanical strength and toughness under dynamic deformations, through slowing poroelastic and viscoelastic relaxations. Moreover, the stretchy VIF network can effectively propagate local stress and strain into a larger region of the cell, deforming more microtubules and actin filaments which interpenetrate and interact with the VIF network. Thus, VIFs significantly enhance the strength and toughness of the cytoplasm, reducing the risk of cell damage during processes involving large deformations.
A cell can’t get by without this incredible network. It’s hard to imagine a primitive protocell surviving without it. Mutations in the genes that make VIF filaments don’t help; they cause significant human diseases. The VIF network design is so good, in fact, that it gives these scientists ideas. Watch for the word design:
Furthermore, the stretchy and hyperelastic VIF network forms a dynamic yet mechanically robust cytoskeleton when combined with the dissipative and quickly recovering cytoskeletal components such as microtubules and actin filaments; such a collaborative design principle may inspire improved engineering of smart materials.
Another Layer of Compartmentalization
One more example will help clinch the point about foresight. Most biology students learn about membrane-bound organelles within cells that keep things together, the nucleus being a prime example. Others include mitochondria, chloroplasts, vacuoles and more. Cells are turning out to be much more organized than that, though, on multiple levels. Here’s some exciting news from MIT about tiny short-term compartments called “droplets” that keep ingredients where they need to be. The nucleus is certainly a phenomenal enclosure for protecting the genetic material and keeping it together;
Yet cells also harbor enclosures that are not membrane-bound and more transient, like oil droplets in water. Over the past two years, these droplets (called “condensates”) have become increasingly recognized as major players in controlling genes. Now, a team led by Whitehead Institute scientists helps expand this emerging picture with the discovery that condensates play a role in splicing, an essential activity that ensures the genetic code is prepared to be translated into protein. The researchers also reveal how a critical piece of cellular machinery moves between different condensates….
“Condensates represent a real paradigm shift in the way molecular biologists think about gene control,” says senior author Richard Young, a member of the Whitehead Institute and professor of biology at MIT. “Now, we’ve added a critical new layer to this thinking that enhances our understanding of splicing as well as the major transcriptional apparatus RNA polymerase II.”
The team found that short-term condensates whimsically called “speckles” are more than just “warehouses” that hold ingredients together needed for transcription of genes and splicing of messenger RNAs. When the cell modifies transcripts with phosphates or other tags, they motivate the speckles into action:
In light of their discoveries, the researchers propose a new view of splicing compartments, where speckles serve primarily as warehouses, storing the thousands of molecules required to support the splicing apparatus when they are not needed. But when splicing is active, the phosphorylated CTD of RNA Pol II serves as an attractant, drawing the necessary splicing materials toward the gene where they are needed and into the splicing condensate.
The team finds this “incredibly exciting” to study, because “It is giving us a whole new way of looking at the world of regulatory biology.”
Cells will never again seem like boring bags of a few squishy parts with names that high school students have to memorize. They truly are appearing as complex as cities, as Michael Denton memorably described them. And we know that the way to build a good city is with lots of foresight. Thank you, Dr. Eberlin, for sending scientists on a fruitful scientific quest to identify purpose in the world around us. It’s everywhere.
Photo: Diatoms, by Prof. Gordon T. Taylor, Stony Brook University [Public domain], via Wikimedia Commons.
