 Intelligent Design
Intelligent Design
Chromosome Dynamics Has Egg-centric Features
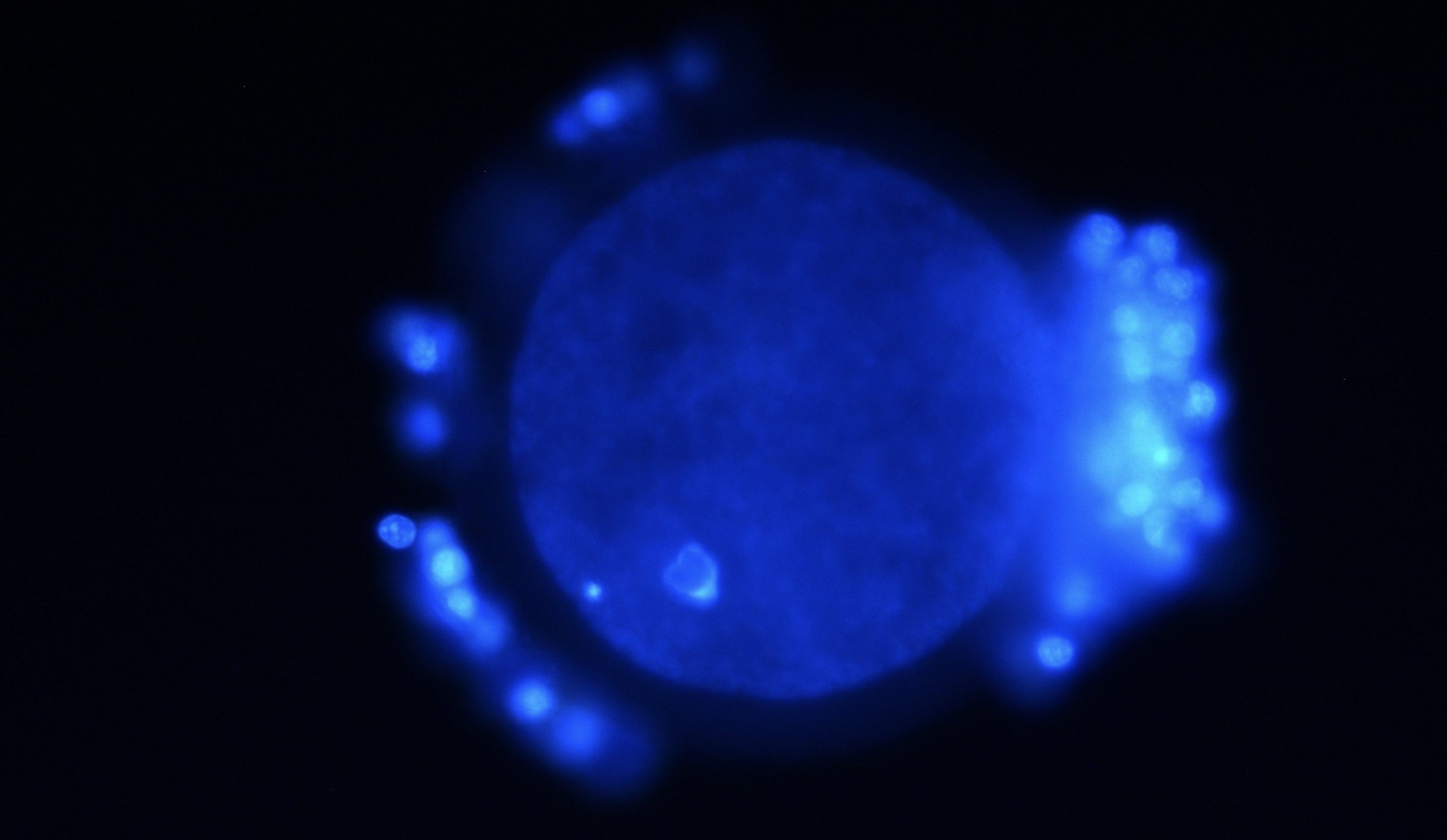
In metazoans ranging from arthropods to vertebrates, chromosomes are not inherited as linear, “beads-on-string” collections of DNA elements. Rather, they are bequeathed (so to speak) as complex 3D/4D plexuses that are unique not only to the gametes wherein they reside, but also to the earliest “pre-phases” of development. (Indeed, such cytological distinctions as those between euchromatin and heterochromatin often emerge in embryogenesis when, say, gastrulation gets underway, as is the case with Drosophila melanogaster.) And it is in the egg cell (oocyte) that we see DNA taking on exceptional roles, such as becoming the cytoskeletal organizer of the meiotic network or spindle, in addition to providing the sites for centromere/kinetochore assembly, both of which in turn govern the formation of the haploid state that precedes fertilization (or in some taxa, parthenogenesis).
To Arrive at an Egg
To begin to see this, we have to take a brief look at certain stages that lead up to a mature egg being produced. Long before an oocyte can meet with its paternal counterpart, its nuclear materials have to be extensively reorganized. For instance, the diploid chromosomal set has to be replicated so that the pre-oocyte becomes (in effect) tetraploid. Then synaptonemal complexes are set up between a diploid subset of (some of) the chromosomal arms, which are proteinaceous (or ribonucleoprotein) scaffoldings that generate DNA-level variability (among other things). By the pairing-up or synapsis of homologous chromosomes (bivalents), some of the chromatin/DNA is subjected to double-stranded breaks and, hence, to the beginnings of meiotic recombination. Remarkably now, I hasten to add, the chiasmata (cross-overs) that are deployed keep the chromosomes “interlocked” for quite some time (from prophase I to anaphase I). Without these chromatin/DNA bridges from one homologue to its partner and vice versa, meiosis will often result in errors in segregation (such as non-disjunction) and transmission that can be unviable for the to-be-formed zygote. And the importance of maintaining such a physical tethering between homologous chromosomes amidst much of meiosis I, is underscored by the fact that there is an inverse correlation between the frequency of cross-overs, on the one hand, and the failure of homologous chromosomes or sister chromatids to properly separate, on the other hand (Baker and Hall 1976).
Exceptional Roles for DNA
This brings us to the point that I want to touch on, namely, that of DNA taking on exceptional roles, such as becoming the cytoskeletal organizer of the meiotic spindle. The bivalents and their chiasmata can determine the events of segregation, but only in the context of dynamic maneuvers that orient properly the homologues to opposite poles. Yet the latter has to occur in the absence of centrosomes — the centriolar-based, microtubule-organizing centers that are necessary for a host of mitotic and non-mitotic processes — as these are eliminated in oogenesis (for reasons unknown; Pimenta-Marques, et al. 2016; Severson, et al. 2016). This “acentrosomal state” is compensated for, however, by the ability of chromatin/DNA to be involved in the nucleation and organizing of the very meiotic network that is required (McKim, Hawley 1995). What this means is that along the female germline track, just prior to and during meiosis I and II, a completely different kind of functionality is superimposed on nucleotide strings — one which goes beyond the erstwhile “coding” versus “non-coding” distinction.
Four Major Events
Without going here into too much detail, based on observations that can be made in vitro, we can note four major events that this functionality governs (see Radford et al., 2017 for most of the following). First, there is the chromatin-/DNA-directed erection of a framework of microtubules around the homologues. This is due in part to the formation of a gradient of Ran-GTP along the chromosomes and the chromosomal passenger complex (at least in Xenopus), which entails a cascade of initial microtubule-organizing steps. Second, there is the effecting of a force that generates the hierarchy of the spindle, with two poles becoming established at either end of the central axis. Third, the chromosomes become dually oriented within the meiotic plexus, as each homologue (with tight centromeric coupling of its sister chromatids) is attached to its correct pole. And fourth, the meiotic spindle is maintained by the cooperative dynamics of multiple microtubule motors, microtubule cross-linkers, regulators of microtubule turnover, and so forth (Costa, Ohkura, 2019). Note that upon reaching metaphase I, the process halts…the spindle is maintained all the while (for days or longer) until meiosis I resumes with egg activation and/or fertilization. Thus, what the oocyte bestows to the zygote-to-be is not naked DNA “replicative strings” that can be scanned by some Turing Universal-machine, but rather an intricate 3D/4D plexus that “bootstraps” its own segregation and transmission.
But this is not to say that because “the chromosomes are more active participants during acentrosomal spindle assembly in oocytes,” it follows that “chromosomes carry information that can direct the meiotic divisions” and that “chromosomes dictate their own behavior” (Radford et al., 2017). Why not? Well, in the words of Robert Rosen (1984) we have to
drastically reconsider what is meant by “genetic information,” and…the role of information in modulating dynamical processes like morphogenesis. The fact is that “information” is never manifested statically (e.g. as in a sequence of nucleotides) or represented in terms of configurational coordinates: it always appears in an active role, in the form of rate constants, or more generally in the determination of a vector field imposed on a manifold of underlying configurational variables. Thus, in general, “information” must be characterized in terms of dynamical interactive capability.
And it is for this reason that I think the acentrosomal meiotic spindle is telling us something profound about how different kinds of functionality are superimposed on the same nucleotide strings, as well as how “information” can be sought in its dynamical events.
Literature Cited
- Baker B.S., Hall J.C. 1976 Meiotic mutants: genetic control of meiotic recombination and chromosome segregation. In: The Genetics and Biology of Drosophila, edited by Ashburner M., Novitski E., pp. 351-434. (eds.). Academic Press; New York.
- Costa M.F.A., Ohkura H. 2019. The molecular architecture of the meiotic spindle is remodeled during metaphase arrest in oocytes. Journal of Cell Biology 218: 2854-2864.s
- McKim K.S., Hawley R.S. 1995. Chromosomal control of meiotic cell division. Science 270: 1595-1601.
- Pimenta-Marques A., Bento I., Lopes C.A., Duarte P., Jana S.C., Bettencourt-Dias M. 2016. A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science 353: aaf4866.
- Radford S.J., Nguyen A.L., Schindler K., McKim K.S. 2017. The chromosomal basis of meiotic acentrosomal spindle assembly and function in oocytes. Chromosoma 126: 351-364.
- Rosen, R. 1984. Genomic control of global features of morphogenesis. In: Modelling of Patterns in Space and Time, edited by W. Jäger et al., pp. 318-330. Springer Verlag, Berlin, Heidelberg.
- Severson A.F., von Dassow G., Bowerman B. 2016. Oocyte meiotic spindle assembly and function. Current Topics in Developmental Biology 116: 65-98.
