 Evolution
Evolution
 Intelligent Design
Intelligent Design
ATP Synthase Gets Smarter
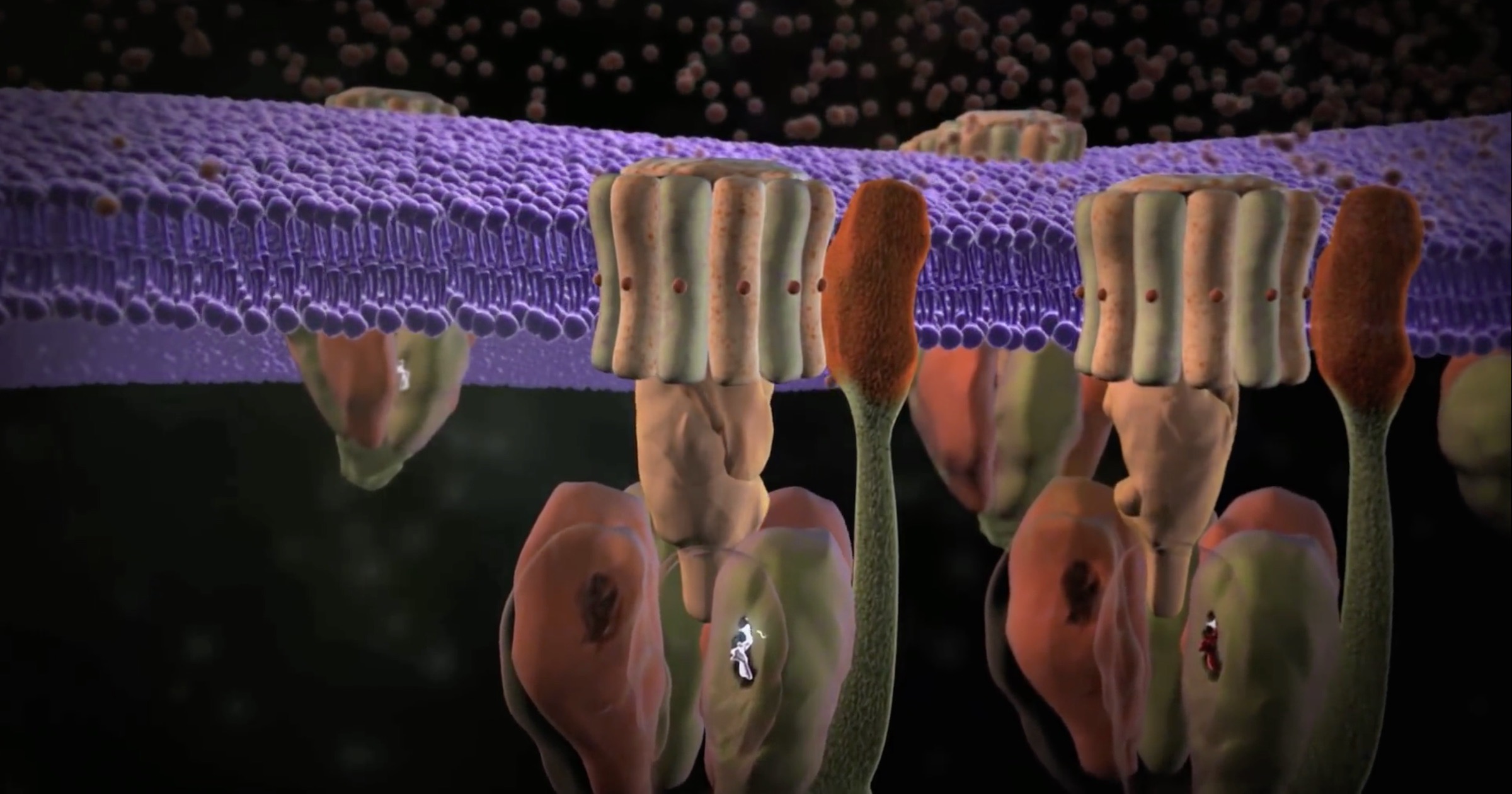
Two new open-access papers about ATP synthase have appeared, one from a team in Germany and one from a team in Japan. Here is a summary of the principal findings.
ATP synthase is an iconic intelligently designed molecular machine because of its rotary engine and its universal distribution. Similar forms of these rotating machines, one-tenth the size of the famous bacterial flagellum, are found in all animal cells, plant cells, bacteria, and archaea. The precision machine raises a huge challenge to all origin-of-life theories: how could a working cell arise without these irreducibly complex molecular machines? They are vital to every organism on earth. I heard a biologist at JPL say that if they stopped working, we would be dead before we hit the floor.
It’s hard not to love this gadget. Discovery Institute’s animation of the machine presents the basics. For eukaryotic ATP synthase, there is a rotor and stator in the FO domain that turns a “crankshaft” to operate the F1 domain. In the F1 domain, three pairs of catalytic units combine ADP (adenosine diphosphate) with phosphate to produce ATP (adenosine triphosphate), the universal energy currency for most cellular processes. A main purpose of the food we eat is to create a flow of ions to operate these machines. ATP synthase is the last machine in a series of complexes in our mitochondria whose collective function is to donate electrons to various intermediates so that protons can be extracted. Protons accumulate between the mitochondrial membranes and flow into ATP synthases. The machines are lined up in pairs along the folds of the mitochondria (cristae) to take advantage of the proton motive force. That flow of protons turns the FO domains like waterwheels, generating three ATP per revolution in the F1 domains, as shown in the animation.
Irreducibly Complex Assembly
Understandably, ever since its rotary mechanism was discovered in 1997, leading to a Nobel Prize, biochemists have been excited about ATP synthase. They continue to pursue every detail, and new methods of super-resolution microscopy are helping them do it. And for some reason, papers about ATP synthase are strangely quiet about Darwinian evolution.
The first paper, in Nature Communications, describes new findings about the assembly of the machine. (Recall that in Unlocking the Mystery of Life, biochemist Scott Minnich noted that the assembly instructions for the bacterial flagellum were more irreducibly complex than the flagellum itself, so this is an important question.) Nina Morgner and four colleagues at Goethe University Frankfurt used “laser induced liquid bead ion desorption mass spectrometry” in their work. They wanted to know if the F1 part would self-assemble in vitro or if chaperones in the cell were required. The short answer is that some of the parts can snap together in vitro, but not the whole complex. Moreover, the self-assembling parts were not as stable as their counterparts in a living cell.
In particular, the α and β subunits in F1 that make up the hexameric head can self-assemble into heterodimers in the presence of nucleotide and Mg2+, but they do not assemble into the complete hexameric head. This surprised the team. Similarly, the γ and ε subunits in the crankshaft-like central stalk could self-assemble but would not link up with the synthase until the hexameric head was in place. The δ subunit, which regulates ATP production, would not combine until everything else was present; see Figure 8 in the paper.
Interestingly, they use the word “interesting” or “interestingly” twenty times! It appears they were having fun figuring this machine out.
An interesting finding was that for the α mutant α[R363Q], stable αβ heterodimers and even higher F1subcomplexes, including one or two heterodimers … can still form, albeit no complex with a complete hexameric head can be detected (Fig. 7d). A similar disturbance was found for incubations of wildtype subunits with AMP-PNP, which might sterically influence the assembly. This shows that even if αβ dimer formation is possible, the assembly of the hexameric head, which has to function via large conformational changes in a very tightly controlled manner, is a precise process, which can be hindered by small disruptions. [Emphasis added.]
Unlike the partly assembled building blocks in vitro, in the cell, everything comes together perfectly. The parts are stable and the machine is stable. This implies the presence of chaperones that guide the parts together. That, in turn, adds to the irreducible complexity; the genetic code must build the chaperones as well as the final product. They conclude that “The entire assembly process follows a choreographed pathway, which allows for efficient construction of the F1 complex.” (Note that we are only speaking of the F1 domain; the FO domain has its own assembly issues.)
Bacteria Do the Spin, Too
The other team, in Japan, wanted to watch the V-type ATP synthase1 in a bacterium to see how it spins. Their paper, also in Nature Communications, tells how they used cryo-electron microscopy to take “snapshots” of 18 intermediates in one cycle of the catalyzing domain. They found a “zipper” and a “ratchet” that keep the machine rotating in the functional direction:
The models reveal that the rotor does not rotate immediately after binding of ATP to the V1. Instead, three events proceed simultaneously with the 120˚ rotation of the shaft: hydrolysis of ATP in ABsemi, zippermovement in ABopen by the binding ATP, and unzipper movement in ABclosed with release of both ADP and Pi. This indicates the unidirectional rotation of V/A-ATPase by a ratchet-like mechanism owing to ATP hydrolysis in ABsemi, rather than the power stroke model proposed previously for F1-ATPase.
It appears that each AB dimer benefits from “cooperativity between the three catalytic sites,” so that as one of them is unzipping phosphate from the ATP, another is zipping the ATP to the binding site and opening the third dimer to accept the ATP for hydrolysis. While these operations occur in discrete 120° rotations, biochemists have noticed dwell times at 80° and 40° substeps, “suggesting the existence of at least one additional catalytic intermediate” within each 120° rotation. Clearly there is more to learn.
These insights were gleaned from studying a thermophilic microbe, Thermus thermophilus (“hot heat-lover”), an “extreme thermophile … aerobic bacterium that can grow at temperatures ranging from 50°C to 82°C (122° to 180° F, see the Journal of Bacteriology). How would evolution build a “primitive” microbe in a hydrothermal vent, as some propose, where its DNA would be subject to degradation by high temperatures? The fact that “hyperthermophiles” like this can maintain their DNA and their molecular machines challenges any notion of “primitive” microbial life.
If evolutionary biologists are interested in these wonders of nature, advocates of intelligent design should be all the more eager to learn about them and to share this information with those unfamiliar with the complexity of all life.
Notes
- V-type or vacuolar ATPases, found in bacteria and in eukaryotic vacuoles, primarily run the reverse reaction, hydrolyzing ATP to generate ions. In eukaryotes, this can acidify vacuoles for breaking down spent proteins. Bacteria use the energy for various functions, but like all other life forms they also have the F-type rotary engines to make ATP. See DifferenceBetween.com for details.
