 Intelligent Design
Intelligent Design
Yes, a “Host of Machines” Are at Work in the Cell
Much as we love the old standbys (the flagellum, cilium, ATP synthase, etc.), we should never assume that the case for intelligent design is restricted to those iconic favorites. In 2002, Jed Macosko claimed in Unlocking the Mystery of Life that there are “a host of machines” at work in the cell — as many as there are functions that the body has to perform. Each machine is a marvel of engineering and precision. Let’s visit a few more of them.
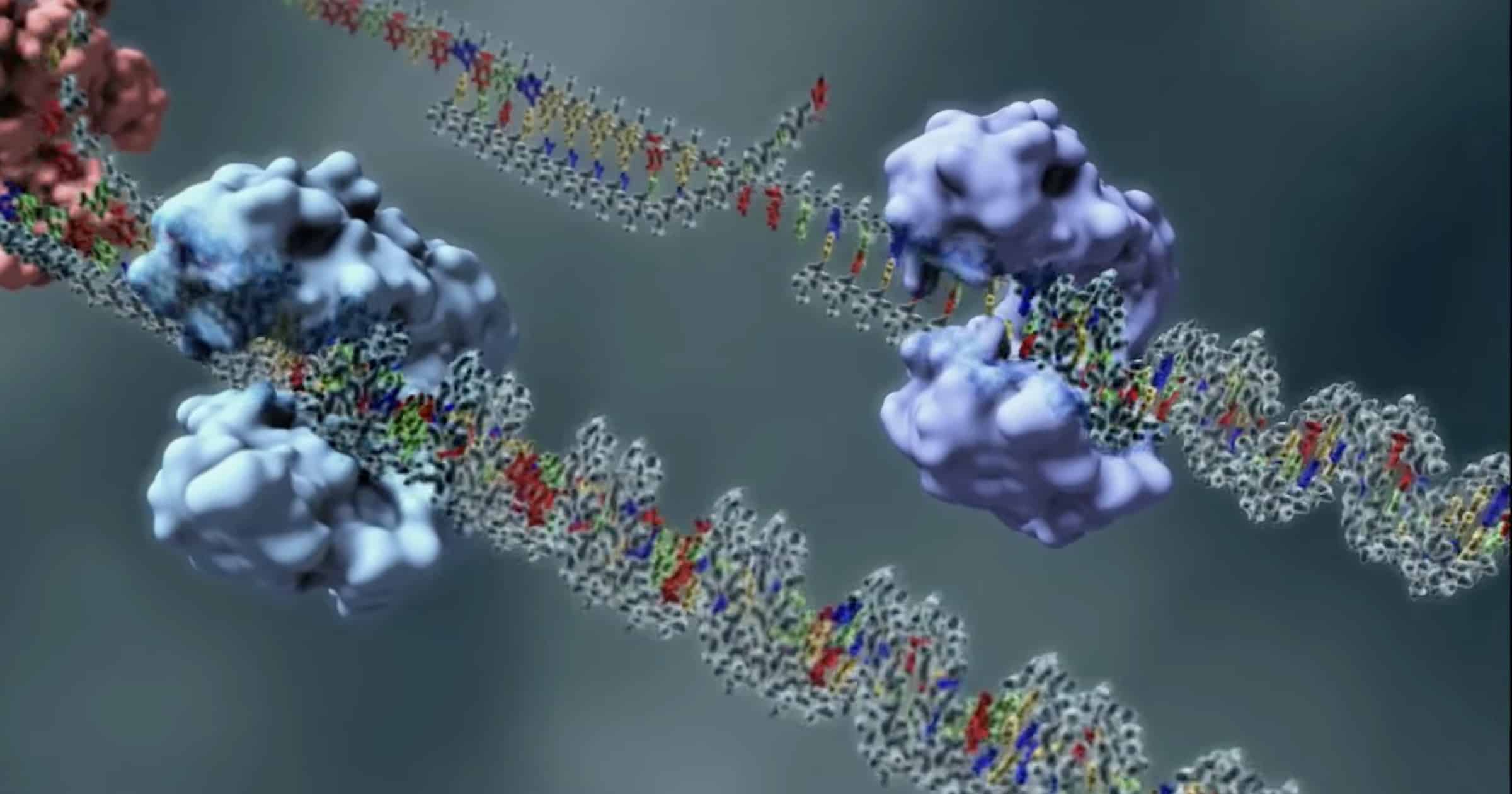
Another Topoisomerase
Discovery Institute’s recent animation of a Type 2 topoisomerase dazzled us with its ability to accurately cut DNA and stitch it back together to fix twisted loops that could send a genome into a tailspin.
The family of topoisomerase machines, though, includes six types just as amazing. Topoisomerase 3β (abbreviated TOP3B) received an ovation this month from Yang et al. in Nature Communications. This one “regulates R-loop dynamics and mRNA translation, which are critical for genome stability, neurodevelopment and normal aging.” TOP3B is a Type 1 topoisomerase that repairs one strand of DNA (see below for a simple video demonstration).
The details are more complex than shown, as this excerpt from the paper demonstrates:
As a Type IA topoisomerase, TOP3B acts by general acid-base catalysis to break and rejoin single-stranded DNA. Passage of a second DNA strand through the transient break permits dissipation of hypernegative DNA supercoiling and catenation/knotting. Additionally, hsTOP3B was recently demonstrated as the human RNA topoisomerase, required for normal neurodevelopment and proposed to be a potential anti-viral target upon RNA virus infection. Here we elucidate the biochemical mechanisms of human TOP3B. We delineate the roles of divalent metal ions, and of a conserved Lysine residue (K10) in the differential catalysis of DNA and RNA. We also demonstrate that three regulatory factors fine-tune the catalytic performance of TOP3B: the TOP3B C-terminal tail, its protein partner TDRD3, and the sequence of its DNA/RNA substrates.
As stated, this machine can precisely break and re-join single strands of DNA to untie knots. Like its Type 2 counterpart that can cut and splice two strands at a time, TOP3B works in a “three-step strategy — DNA cleavage, strand passage, DNA rejoining.” It requires three fine-tuned regulatory cofactors to work. And it is required for normal neurodevelopment.
One other surprise is worth noting: the need for metal ions to cut the nucleic acids: “addition of Mg2+ and to a greater extent Mn2+ stimulated DNA binding and cleavage by TOP3B.” Is it fair to say that this machine has metal cutting blades?
Another Dart Gun
We’ve learned about the Type 3 Secretion System and the Type 4 Secretion System in bacteria that shoot molecules outside their membranes, but there’s another we haven’t considered for a while: Type 6. An Evolution News article in 2015 discussed its firing mechanism. Now, Lin Lin and colleagues at the University of Basel, Switzerland, have published an open-access paper in the EMBO Journal describing T6SS assembly. The machine looks like a cannon or bazooka in the figures. Remarkably, this “large nanomachine that can deliver toxins directly across membranes of proximal target cells” appears to assemble rapidly on contact when the bacterium encounters a rival in “interbacterial competition.”
We identified a class of diverse, previously uncharacterized, periplasmic proteins required for this dynamic localization of T6SS to cell–cell contact (TslA). This precise localization is also dependent on the outer membrane porin OmpA. Our analysis links transmembrane communication to accurate timing and localization of T6SS assembly as well as uncovers a pathway allowing bacterial cells to respond to cell–cell contact during interbacterial competition.
This implies that the components are stored locally throughout the inner membrane to avoid the cost of maintaining an arsenal all over the cell. It also implies contact sensors to activate the assembly — which the authors found.
In this work, we reveal a mechanism of localization of a complex nanomachine in response to contact with another cell. Such precise positioning likely requires temporal and spatial coordination of many regulatory proteins at the membrane. Since many T6SS accessory proteins remain uncharacterized, we propose that some of these proteins might be required for spatiotemporal regulation of T6SS assembly. We expect that additional mechanisms of dynamic localization of T6SS will be identified in more bacteria and shown to play an important role in their pathogenesis or ecology.
The authors use strategic mission terms like “attack” in their belief that the T6SS “likely evolved to allow efficient killing of target cells.” But we shouldn’t take the warfare motif too seriously. They’re just bacteria. Maybe the colony is programmed to regulate its surroundings this way. They expect that higher resolution imaging will “unravel additional intricate mechanisms of T6SS targeting.”
Nitrogenase Black Box
One of the most important enzymes in all nature, nitrogenase has defied analysis. This multi-part machine with metals in its core is the only enzyme that can split the tough triple bonds of atmospheric nitrogen. The enzyme is only found in bacteria that form symbiotic relationships with the roots of some plants. Biophysicists have tried for years to open the black box of nitrogenase, but its conformational changes are fleeting and difficult to capture.
Reduced forms of nitrogen are essential for the biosynthesis of amino acids and nucleic acids as well as the production of fertilizers and many commodity chemicals. As the only enzyme capable of nitrogen fixation, nitrogenase catalyzes the eight-electron reduction of atmospheric nitrogen (N2) and protons (H+) into ammonia(NH3) and hydrogen (H2) (Fig. 1A). Nitrogenase is a two-component enzyme, which, in its most common form, consists of the iron protein FeP (a γ2 homodimer) and the molybdenum-iron protein MoFeP (an α2β2 heterotetramer) (Fig. 1B) (4, 5). Nitrogenase is distinct from most redox enzymes in its requirement for adenosine triphosphate (ATP) hydrolysis to enable the successive transfer of electrons and protons for substrate reduction
Hannah L. Rutledge at UC San Diego (Science) used cryo-electron microscopy and abundant ATP to capture a few more frames in the “movie” of this remarkable and unique machine. Based on the states they were able to capture, they think they see how the game is being played:
Given the asymmetry between the FeMoco’s in the two αβ halves, it is tempting to propose a “ping-pong”–type mechanism in which the cofactors proceed through each of the eight catalytic steps in an alternating fashion. This scenario would assign a dual role to FeP: (i) to deliver an electron to one αβ subunit of MoFeP and (ii) to suppress FeP binding to the opposite αβ subunit while priming it for catalytic transformations through long-distance activation of electron, H+, and/or substrate access pathways to the distal FeMoco. Establishing whether such a mechanism is operative will require future studies, but our current work illustrates that it is possible to characterize nitrogenase under turnover conditions by cryo-EM at near-atomic resolution, representing a critical step toward understanding the mechanism of this enigmatic enzyme in full structural detail.
When that day comes, it could revolutionize agriculture. An article from Tsinghua University Press (Phys.org) says that “major breakthroughs are still badly needed” to replace the energy-intensive Haber-Bosch process which “is disrupting the planet’s nitrogen cycle, warming the globe, and potentially risking the health of millions.” Imitating the way cells do it might allow scientists to fix nitrogen at ambient temperatures. Nitrogenase is a molecular machine to watch!
Rubisco Geometry
Another vital enzyme for the biosphere is Rubisco, which fixes carbon for conversion to sugars. Emily Reeves wrote about how it works last year, so readers are referred to that article for background. A new open-access paper in Nature Communications by Lauren Ann Metskas et al. adds some interesting details about its geometry: it forms a lattice inside tiny geometric domes.
This arrangement preserves freedom of motion and accessibility around the Rubisco active site and the binding sites for two other carboxysome proteins, CsoSCA (a carbonic anhydrase) and the disordered CsoS2, even at Rubisco concentrations exceeding 800 μM. This characterization of Rubisco cargo inside the α-carboxysome provides insight into the balance between order and disorder in microcompartment organization.
The research team points out that many prokaryotes segregate their machines into microcompartments like this, contrary to earlier assumptions that the enzymes just float around in the interior. Like the cell’s outer membrane, a proteinaceous shell “selectively restricts passage of key intermediates and improves on-target catalysis.”
Interest in “microcompartment bioengineering” has grown in recent years, the authors say. The paper shows electron micrographs of some of these remarkable geometric compartments with the Rubisco enzymes neatly packed inside but “surprisingly” not in contact with the shell. “The six-fold lattice may therefore be a packing mechanism to preserve function at concentrations that could otherwise crystallize or sterically impede function.” That makes design sense.
Microcompartment bioengineering is a growing field, and this polymerization-based mechanism for efficient, functional, and controlled packing of enzymes may be useful for future discovery and designs in both CBs [carboxysomes] and other microcompartments.
Once again, observation and expectation of design from an engineering perspective is proving fruitful in opening the black boxes of the cell and peering at the watch-like moving parts inside. The future looks bright for nanobiotechnology, and with it, increases in global human flourishing. Intelligent design is proving to be “useful for future discovery and designs.”
