 Intelligent Design
Intelligent Design
DNA May Be “Junk” at One Level But of Utmost Significance at Another
In a previous article I mentioned that chromosomes can and do have roles that go far beyond the so-called “Central Dogma.” Nowhere is this point better demonstrated, it would seem, than in the phenomenon of meiosis or “reductional division,” which generates a haploid gamete from a diploid germ cell by means of an intricate series of developmental events. Indeed, it is during the latter stages of the production of an animal oocyte that many functionalities of what some disparage as “junk DNA” take center stage.
Recall that in almost-mature eggs from arthropods to vertebrates the meiotic networks are acentriolar (or acentrosomal or anastral), because the chromosomes to be divided eventually both recruit and sequester tubulin. In this way they are involved in organizing the bipolarity of the required spindle (Matthies et al. 1996, Theurkauf and Hawley 1992). The latter is composed (at least in part) of anti-parallel microtubules in the central region, which interact on the lateral margins with the chromosomal masses by the time of the metaphase-I arrest. In addition to this, microtubules that run from pole to pole come to intersect at the “equator” where the chromatin is positioned (Jang et al. 2005). We have here, then, an instance of a bipolarized cytoskeleton of which DNA elements (both “coding” and “non-coding”) are warp and weft.
“Over and Above”
But this is only part of the process. Chromosomes that are about to undergo meiosis (or mitosis) have a distinct ensemble of functions that are grouped and harnessed ever so intricately into kinetochores, which are the “levers” of their maneuverings. These are “over and above” (indeed, epi– to) nucleotides have long been observed to be peculiar in their qualities. For one thing, they are dynamic platforms that have roles that are phase- or stage-specific. In the case of metaphase I, for example, those of one set of sister chromatids have to be in cohesion (or be paired) and jointly linked to one pole, while those of the other set of sister chromatids have to likewise be in cohesion (or be paired) but jointly linked to the opposite pole (Hughes et al., 2018), as I have depicted here in an oversimplified way:
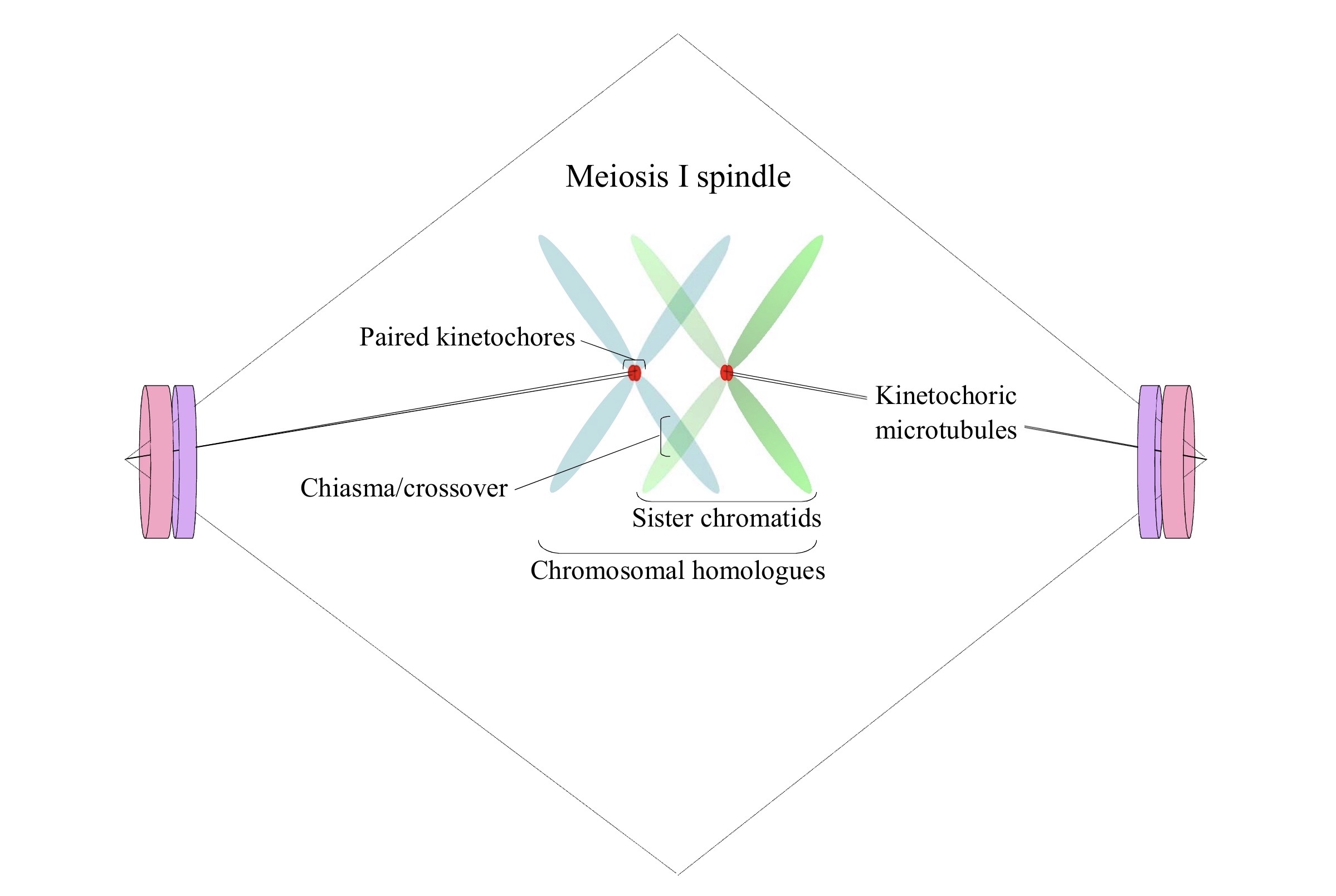
For another, as the chromosomal homologues are aligned at the “equator” of the meiosis I nexus, with chiasmata locking together the arms that have undergone crossovers, both paired-kinetochoric and spindle symmetry has to be maintained overall by kinesins and various other proteins (Radford et al. 2017). Then again, the chromosomal passenger complex is necessary for the generation of this spindle, and certain components of it such as Aurora B kinase and Incenp are localized in a centralizes ring around the chromatin/DNA (Jang et al. 2005; Radford et al. 2012). Leaving much out (see Hughes et al., 2018), what is to be emphasized is that there are specialized chromatin regions, kinetochores, which are fundamental to the proper orientation and segregation of the chromosomal sets during meiosis I.
Kilobases to Megabases
Now, the lowermost tier of a kinetochore is generated on (and confluent with) a centromere or centromeric locus, and what sets the latter apart on any chromosome is its distinct histone composition. Yet when one turns to the basic constitution of such in terms of the DNA elements that underlie it, all that one can find is junk…kilobases to megabases of junk. Let the model organism Drosophila melanogaster underscore this fact for us. First, all known centromeric loci in this species have nucleotide orderings that are “typographically” unique. This is not because they lack repetitive sequences but rather because they do — each is composed of “complex islands” of dispersed and scrambled retroelements (especially those of the G2/Jockey-3 family of Long Interspersed Nuclear Elements or LINEs) that are concatenated with a host other “junk” remnants, such as DNA transposons (Chang et al. 2019). Second, the same are flanked by tandem repeats such as (AAGAG)n, (AAGAT)n, (AATAG)n, and/or others that have longer monomer units (dodeca and Prodsat). Third, there is no motif that is centromere-specific for all the sequence families are represented elsewhere in the genome of this taxon. Fourth, the extent of these loci can vary manyfold. (See Chang et al. 2019 for these and other details.) This adds credence to the notion that these regions are “epigenomic” as opposed “genomic.”
What is of importance, though, is that we keep two pieces of evidence in mind when we take in such details. On the one hand, the kinetochores/centromeres are of the utmost significance to the D. melanogaster female meiosis I network, for they are foundational to the generation of a viable zygote. On the other hand, though, of the four chromosomal homologues (namely, those of the 2nd, 3rd, 4th, and X; normally no Y chromosome is present) that are present in such a spindle, their kinetochores/centromeres are layered on these segments:
- Those of the 2nd are named Capri and each consists of an approximately 1,800 base pair region, which is enriched in G2/Jockey-3 and flanked by the (AAGAG)n and (AATAG)n units.
- Those of the 3rd are named Giglio and each consists of an approximately 68,500 base pair region, which includes 240 copies of a variant of the ribosomal-RNA Intergenic Spacer along with interspersed blocks of G2/Jockey-3 and DNA transposons; it is flanked by the dodeca and Prodsat units.
- Those of the 4th are named Lampedusa and each consists of an approximately 42,800 base pair region, which is enriched in G2/Jockey-3 and other non-retroviral-like transposable elements; it is flanked by the (AAGAT)n units.
- Finally, those of the X are named Maupiti and each consists of an approximately 70,000 base pair region, with long stretches of G2/Jockey-3 plus other complex, divergent repeats; it is flanked by a short stretch of (AAGAT)n units on one of its sides but overall it is surrounded by the (AAGAG)n units.
A Top-Down Causal Process
What these two pieces of evidence suggest, then, is that what may be “junk” at one level may be of the utmost significance at another level. This happens I think by way of a top-down causal process that Paul Davies (2012) has alluded to, explainable in part by the augmentation of
the normal terms referring to local forces contained in the Hamiltonian for chromatin with additional (presumably small) non-local terms representing functional (i.e. semantic or contextual) information. By coupling the mechanical and informational dynamics in this manner, the dynamical laws describing chromatin behaviour would become time-dependent and change according to the informational state of the system. Information would then possess direct, albeit subtle, traction over matter and permit epigenetic control to be exercised directly on the chromatin itself.
Such would involve an “explicit coupling between dynamical laws and information-rich states, thus endowing higher level entities, such as contextual information, with direct causal efficacy on matter alongside intermolecular forces.”
If we take such “contextual information” to be resident at all tiers of the meiosis I network, from the spindle as whole to (say) each and every centromeric locus, then “with direct causal efficacy” retroelement and satellite DNAs can be endowed with information-bearing states. Of course, “such a proposal represents a decisive break with the normal formulation of the theory of dynamical systems,” as Davies notes, which means that “theories of this sort remain largely unexplored.” But explore it we should…and especially where cytology meets “junk” DNA.
Literature
Chang C.-H., Chavan A., Palladino J., et alia. 2019. Islands of retroelements are major components of Drosophila centromeres. PLoS Biology 17: e3000241.
Davies P.C.W. 2012. The epigenome and top-down causation. Interface Focus 2: 42-48.
Hughes S.E., Miller D.E., Miller A.L., Hawley R.S. 2018. Female meiosis: synapsis, recombination, and segregation in Drosophila melanogaster. Genetics 208: 875-908.
Jang, J.K., Rahman T., McKim, K.S. 2005. The kinesinlike protein Subito contributes to central spindle assembly and organization of the meiotic spindle in Drosophila oocytes. Molecular Biology of the Cell 16: 4684-4694.
Matthies H.J., McDonald H.B., Goldstein L.S., Theurkauf W.E. 1996. Anastral meiotic spindle morphogenesis: role of the nonclaret disjunctional kinesin-like protein. Journal of Cell Biology 134: 455-464.
Radford S.J., Jang J.K., McKim K.S. 2012. The chromosomal passenger complex is required for meiotic acentrosomal spindle assembly and chromosome biorientation. Genetics 192: 417-429.
Radford S.J., Go A.M., McKim K.S. 2017. Cooperation between kinesin motors promotes spindle symmetry and chromosome organization in oocytes. Genetics 205: 517-527.
Theurkauf W. E., Hawley R.S. 1992 Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. Journal of Cell Biology 116: 1167-1180.
