 Intelligent Design
Intelligent Design
More Information Found in DNA: The Shape Code
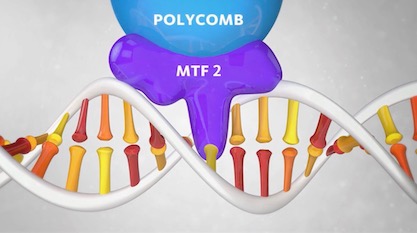
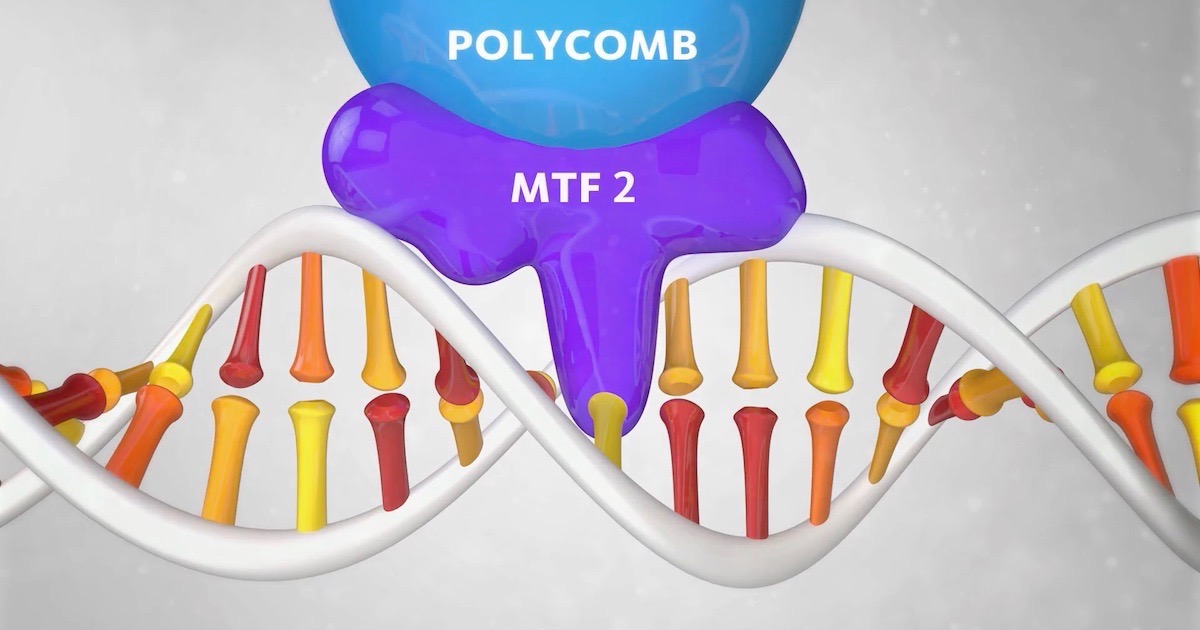
We may have yet another code to add to Jonathan Wells’s growing list of information systems in the cell that challenge the Central Dogma. A new discovery hints at a “shape code” in the double helix.
Can the shape of the DNA double helix affect its behavior? Researchers at Radboud University in the Netherlands made “a remarkable discovery” about a protein named Polycomb that binds to DNA. They noticed that it would not bind unless the helix relaxed its twist slightly at the binding site. They concluded, “The shape of the DNA helix proves to be as important as its sequence.”
The mechanism of DNA binding of the well-studied protein Polycomb, which is vital for cell division and embryogenesis, has finally been deciphered. A remarkable discovery, as it proves that the shape of DNA is at least as important for where the protein binds in the DNA as the DNA sequence. The role of the shape of DNA had not been demonstrated so clearly. Researchers at Radboud University will publish their findings on May 28th in the scientific journal Nature Genetics. [Emphasis added.]
A 13-second animation shows how this works. The docking protein MTF2, which ferries Polycomb, needs the DNA to un-twist in order to bind to the site, where Polycomb will switch off specific genes. “MTF2 only recognises the binding spot on the DNA if the helix is in a relatively unwound state,” the caption says.
The scientists say this is a new way to look at DNA. What they found may be a general feature of how it functions — not just by sequence, but by shape.
Besides the classical interpretation of the code (the ‘sequence of letters’) in DNA defining its function, it has been known for several years that the helix shape of DNA may also play a role. “We are currently able to read what is written in the human genome, but understanding the mechanisms is not an easy task,” says Gert Jan Veenstra, Professor of Molecular Developmental Biology, one of the researchers involved in this study. “The concept that helix shape is also involved in how DNA functions, is an interesting new way of perceiving DNA. It could lead to understanding its functioning in general and of the way in which proteins can bind to DNA in certain places.”
The impact of shape on protein function is well known, but the role of DNA shape is relatively less understood.
Give It a Twist
While not entirely new, the concept of a shape code has been more clearly demonstrated in this instance than ever before. This is one of several discoveries about Polycomb mentioned in the paper in Nature Genetics. One subsection in the paper states, “DNA sequence and helical shape dictate MTF2 binding.” At least in this instance, the sequence and the shape play complementary roles. This means that the amount of twist in the double helix is an integral part of the information in DNA.
How did they determine that the shape was a critical factor for the binding site in DNA? They identified binding sites and their flanking regions, then they created “bait” motifs containing mutations in those regions.
To rule out effects due to the sequence surrounding the motif or to the mutation itself, we repeated the experiment using baits with different flanking sequences and different point mutations in the motif…. Notably, PRC2 recruitment was strongly reduced by DNA methylation….
Additional tests narrowed down the shape of the helix as the critical factor where MTF2 would bind.
Not the Sequence but the Shape
The flanking regions controlled the tightness of the helical twist at those points. It wasn’t the sequence; it was the shape.
Specifically, the central unmethylated CpG dinucleotide was critical but not sufficient for binding, as shown by the effect of flanking mutations that also affect the helical structure of the bait. Moreover, the mutations that most severely reduced MTF2 binding cause helical shape perturbations that lie outside the average shape profile of positive-scoring k-mers, while the least perturbing one almost perfectly mirrored the shape of the wild-type bait (Fig. 5b), lending further support to a role of DNA helical shape in MTF2 binding to DNA.
To clinch their detective work, they made some predictions and tested them.
To further investigate the role of DNA shape in determining MTF2 binding sites, we tested whether we could predict MTF2 bound regions using only shape information. We predicted the DNA shape of all the GCG trinucleotides in MTF2 peak summits and used machine learning to classify them against nucleotide-composition-matched controls…. The algorithm was able to identify differences between MTF2-bound vs. unbound unmethylated islands on the basis of helical shape alone….
Taken together, these analyses document the sequence and DNA helical shape properties of MTF2 binding and their role in PRC2 recruitment….
How specific is the shape information? The “bait” sequences demonstrated a “highly specific” binding of the components needed for Polycomb function. In Figure 6 of the open-access paper, the differences between “shape qualifying” and “non-qualifying” amounts of helical twist appear slight, as if a threshold of untwist is necessary to get MTF2 to bind. Flanking regions containing CGC sequences were required to untwist the helix; “Among top-scoring motifs, we identified TGCGCAAA as the most strongly enriched motif in both vertebrate species,” they say. So here, sections of DNA control the twist but not the RNA transcripts, with methylation of the bases also implicated in the effects. These are epigenetic sources of information, not genetic (sequence-based) codes.
The authors used several tests to rule out sequence information as the cause of binding success or failure. They also noted that “shape features might provide directionality to the binding site,” suggesting an additional functional role for the amount of twist. The authors did not use the term “shape code” as we do here, but what else would you call specific flanking sequences and methylation patterns that change the twist or untwist, controlling the binding of transcription factors?
What a Surprise
Recognition of this new source of epigenetic information suggests a “new way of practicing biology,” they say.
The discovery of the mechanisms involved of DNA binding by Polycomb is one of the first concrete examples in which the shape of DNA plays a more important role for the protein’s functioning than the code contained in the DNA. It turns out that the protein can only bind to the DNA helix if the latter is relatively unwound. Veenstra: “Because the DNA-binding protein does not bind to a specific sequence in the DNA, it was difficult to find the working mechanism of the protein using regular research methods.” This mechanism had been actively sought by many people in the field for the past twenty years.
What a surprise, they don’t have much to say about evolution. The only mention in the paper concerned pressure against it. The caption in Figure 2 says:
The lysine rich region (K-rich) of MTF2 is well conserved among vertebrates compared to the other PCL proteins in mouse (a), suggesting an evolutionary pressure against mutation in this area.
What we appear to be observing in this case is another layer of information riding on top of the sequence, governing how the genetic sequences are regulated epigenetically. A design-oriented approach may yet uncover additional instances where the shape of the double helix governs how it is read.
Image source: Radboud University, via YouTube (screen shot).
