 Evolution
Evolution
 Intelligent Design
Intelligent Design
No. 2 Story of 2023: Intelligent Design Passes the Dawkins Test

Editor’s note: Welcome to an Evolution News tradition: a countdown of our Top 10 favorite stories of the past year, concluding on New Year’s Day. Our staff are enjoying the holidays, as we hope that you are, too! Help keep the daily voice of intelligent design going strong. Please give whatever you can to support the Center for Science and Culture before the end of the year!
The following was originally published on May 15, 2023.
Richard Dawkins is famed as an evolutionary biologist and as an aggressive advocate for atheism. He has made some strong statements about the nonexistence of a designer behind life, including in his bestselling book The God Delusion. In light of his position, here is a question someone ought to put to him. Scientists often work by making predictions. As evidence comes in, the prediction can be shown to be true or false. This may have serious consequences. For his part, Dawkins has repeatedly described a test that at the time he thought proved evolution. Now, however, taking into account accumulating scientific evidence, the same test demands a conclusion of intelligent design (ID). Will he accept that conclusion and come out as an ID proponent?
The test that Dawkins has formulated goes this way: He says that if Darwinian evolution is correct, every gene in a group of organisms will give “approximately the same tree of life.” If ID is correct, on the other hand, the designer could have “picked and chosen” the “best proteins for the job” in each organism. In that case, he says, genes would not all give the same tree of genetic resemblances.
Dawkins offered this test in his 2009 book, The Greatest Show on Earth: The Evidence for Evolution. He described it as “extremely powerful evidence for evolution.” At the time, he apparently believed that each gene really did give approximately the same tree. He wrote:
Comparative DNA (or protein) evidence can be used to decide — on the evolutionary assumption — which pairs of animals are closer cousins than which others. What turns this into extremely powerful evidence for evolution is that you can construct a tree of genetic resemblances separately for each gene in turn. And the important result is that every gene delivers approximately the same tree of life. Once again, this is exactly what you would expect if you were dealing with a true family tree. It is not what you would expect if a designer had surveyed the whole animal kingdom and picked and chosen — or “borrowed” — the best proteins for the job, wherever in the animal kingdom they might be found. [Emphasis added.]
pp. 321-322
He’s Said It Other Times
That’s not the only time Dawkins has made this prediction. He said it in an interview given in 2009:
The single most convincing fact or observation you could point to would be the pattern of resemblances that you see when you compare the genes … of any pair of animals you like … and then plot out the resemblances and they form a perfect hierarchy, a perfect family tree. And the only alternative to it being a family tree is that the intelligent designer deliberately set out to deceive us in the most underhanded and devious manner.
He said it again in a 2010 interview:
You get the same family tree. You also get the same family tree if you take genes that are no longer functional, that are just vestigial, they’re not doing anything. … This is overwhelmingly strong evidence. The only way you could get out of saying that that proves that evolution is true is by saying that the intelligent designer, God, deliberately set out to lie to us, deliberately set out to deceive us.
It’s a simple test. Dawkins sets up two competing predictions: one for evolution and one for intelligent design. He claims that evolution predicts “perfect” congruency among different representations of the tree of life — evidence that is so “powerful” he believes it “proves that evolution is true.” On the other hand, he gives incongruency or conflicts among different gene-based trees as a prediction of the “alternative” to evolution, namely intelligent design.
What Do the Data Show?
Now, years later, scientists have sequenced a great number of whole genomes. And as a consequence, they know that Dawkins was wrong. Every gene does not deliver “approximately the same tree of life.”
This is an issue that I and others have written about before (see here, for example), but I thought of it again recently when I watched the inaugural lecture of a professor of evolutionary genomics at Queen Mary University of London. The lecture is intriguingly titled “Trees of Life: Do They Exist?”
The professor, Dr. Richard Buggs, mentions that scientists doing phylogenetic studies normally assume the existence of the Darwinian tree of life. It is quite rare in the literature to find someone actually seeking to prove its existence. He points to Dawkins’s statements in The Greatest Show on Earth as one of these rare examples, along with a Nature paper published in 1982 that Dawkins references. In the lecture, Buggs quotes Dawkins, who says “every gene delivers approximately the same tree of life.” Buggs then comments:
Many of you who work, like me, with sequence data every day, probably winced a little bit when you heard me read that out because, you know, it is actually not the case.
Indeed, Buggs’s own research led to a paper he co-authored in Nature Ecology & Evolution — “Convergent molecular evolution among ash species resistant to the emerald ash borer” — where different genes led to widely different gene trees among closely related species of ash trees. In his lecture he notes that yes, the paper produced a “consensus” tree, but that doesn’t mean the genes gave the same tree. Instead, it was “the best tree that we could get out of all the very, very divergent gene trees.”
That example might be dismissed by Dawkins because it is looking only at species within a genus, and the species might still be capable of crossing with each other. But Buggs points out that different genes are found to give different trees even when the species being studied are very different. He gives the example of a paper that studied the relationships of eudicots (a group of flowering plants that have two seed leaves). This is penetrating deep into the tree of life. He shows a diagram from the paper revealing the “discordance” of various gene-based trees:
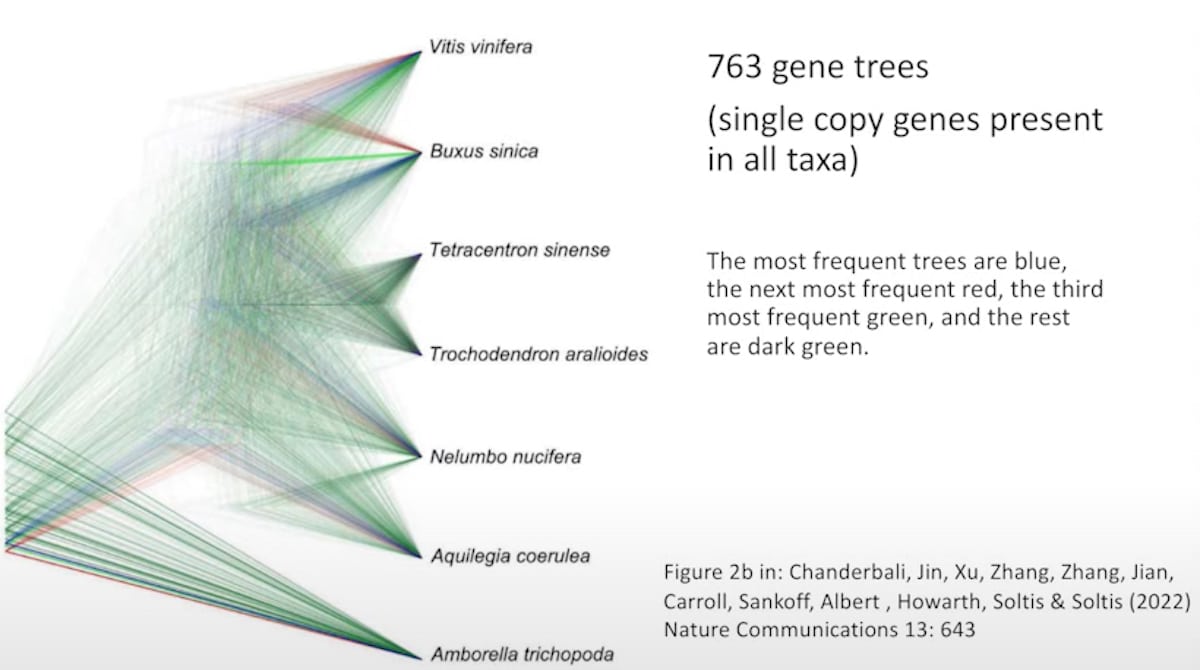
Dr. Buggs then comments:
So according to Richard Dawkins’s test of the Darwinian Tree of Life, we don’t have much support for it, do we? It looks more like the hypothesis that he set up for an intelligent designer.
On its own terms, the Dawkins test for evolution has come up for ID.
So where does Richard Dawkins now stand on this issue? Does he still think that this is a powerful test for ID versus Darwinian evolution? Will he abandon the test, or is he now persuaded by ID? Or has he now changed his mind about how evolution works?
Many Other Recent Papers
It isn’t just Dr. Buggs who is saying this. There are many other examples from other recently published papers showing incongruence among trees of life (all emphases added):
- Dominik Schrempf and Gergely Szöllősi (2020). The Sources of Phylogenetic Conflicts. In Scornavacca, C., Delsuc, F., and Galtier, N., editors, Phylogenetics in the Genomic Era, chapter No. 3.1, pp. 3.1:103.1:23 (2021).
This paper finds that a “conflicting phylogenetic signal between genes is commonplace.”
- Richard H. Adams, Todd A. Castoe, and Michael DeGiorgio, “PhyloWGA: chromosome-aware phylogenetic interrogation of whole genome alignments,” Bioinformatics, 37(13), 1923–1925 (2021).
Says this paper: “An immediate challenge is to address the pervasive phylogenetic conflict observed in whole genome data.”
- Caroline Parins-Fukuchia, Gregory W. Stull, and Stephen A. Smith, “Phylogenomic conflict coincides with rapid morphological innovation,” Proceedings of the National Academy of Sciences (PNAS), 118, No. 19: e2023058118 (2021).
This paper notes that, “Phylogenomic conflict, where gene trees disagree about species tree resolution, is common across genomes and throughout the Tree of Life.”
- Gonzalo Giribet, “Genomics and the animal tree of life: conflicts and future prospects,” Zoologica Scripta, 45: s1, pp 14-21 (2016).
Similarly, this paper states: “A ‘new kid on the block’, phylogenomics, is adding another type of controversy never seen before in molecular phylogenetics: highly supported contradictory results.”
- Nosenko et al., “Deep metazoan phylogeny: When different genes tell different stories,” Molecular Phylogenetics and Evolution, 67: 223-233 (2013).
This paper reviews studies examining the relationships of animals and finds many examples of conflicts:
To unravel the causes for the patterns of extreme inconsistencies at the base of the metazoan tree of life, we constructed a novel supermatrix containing 122 genes, enriched with non-bilaterian taxa. … Different gene matrices tell different stories … The lack of resolution for the deep nodes in this tree reflects major conflicts between the previously published metazoan phylogenies … the best-fitting model left the relative positions of the Bilateria, Coelenterata, and Placozoa–Porifera clades unresolved. … The multiple conflicting metazoan phylogenies presented here and in previous publications have one feature in common: they have long terminal and short internal branches. … To summarize, this study generated three incongruent, yet strongly supported tree topologies…
- Reddy et al., “Why Do Phylogenomic Data Sets Yield Conflicting Trees? Data Type Influences the Avian Tree of Life more than Taxon Sampling,” Systematic Biology, 66(4): 857-879 (2017).
This paper also notes that scientists hoped that by sampling large amounts of data, they would resolve conflicts among trees. But the hope was dashed, especially within the class of birds:
Phylogenomics, the use of large-scale data matrices in phylogenetic analyses, has been viewed as the ultimate solution to the problem of resolving difficult nodes in the tree of life. However, it has become clear that analyses of these large genomic data sets can also result in conflict in estimates of phylogeny. … [P]hylogenomics seemed poised to fulfill this promise to resolve the tree of life. However, analyses of large data matrices have sometimes yielded incongruent topologies, emphasizing that data collection alone is not sufficient to reach this goal.
- Cédric Blais and John M. Archibald, “The past, present and future of the tree of life,” Current Biology, 31: R311-R329 (2021).
This review of the debate between proponents and critics of the “tree of life” hypothesis acknowledges how frequently gene-based trees can conflict, though it postulates that some vertical signal of descent is still present even if it is obscured by processes such as horizontal gene transfer:
Some twenty years ago, the foundations of the tree of life were shaken by the realization that prokaryotic genomes are comprised of genes with different evolutionary histories. … The subsequent explosion of whole-genome sequencing in the late 1990s and the rise of comparative genomics was expected to validate and solidify Woese’s tree. These hopes were short-lived. Phylogenetics soon showed that different genes within the same genome could yield very different tree topologies, and even closely related organisms were found to differ substantially in gene content. A new evolutionary force was invoked: lateral gene transfer, the exchange of genetic material between different species. …
[…]
Resolving a statistical tree of life that unites all extant species may still be possible, contrary to the expectation that only localized tree patterns would survive pervasive lateral gene transfer. But it no longer corresponds to a complete, all-encompassing representation of the genetic history of organisms, as it does not follow any single gene’s history. If the tree of life still stands, it does so in a qualified sense. …The continued use of the tree of life for classification is thus as much a reflection of its practical convenience and historical and cultural inertia as it is a commitment to natural classification.
Extracting phylogenetic signal from genomic data can be difficult, and much of the evidence for ancient relationships is inconclusive at best. Phylogenetic methods designed to build a tree will do so whether it is the best fit with the underlying data or not. Networks, on the other hand, capture incongruences in genomic data without imposing an interpretation.
- Juli Berwald, “Why evolution is not a tree of life but a fuzzy network,” Aeon (2022).
This article quotes geneticist Rasmus Nielsen of UC Berkeley: “That whole abstraction of evolution as being a tree, we always knew was a little inadequate … But now we know it’s really inadequate.”
Nielsen continued: “‘I think that process of splitting up and merging back together again, and getting a bit of DNA from here to there, that’s happening all the time, in all of the tree of life,’ Nielsen said. ‘And it’s really changing how we’re thinking about it, that it really is a network of life, not a tree of life.’”
The article thus describes the history of life as a reticulated pattern rather than a tree: “The hypothesis of reticulate evolution is that species are not as isolated from each other as Haeckel’s branching trees propose. Instead, species both diverge and merge together. The tree of life doesn’t look like a tree so much as the reticulated pattern of a python’s skin.”
This phenomenon of genes appearing in locations not expected by traditional vertical common descent is found very frequently, for “Roving genes have been found in every branch of the tree of life where geneticists have looked.” The article asks “If species don’t rest neatly on the ends of tree branches, what does that mean for Haeckel’s model of the evolutionary tree? Should we throw it out?”
A paper examined studies of fish relationships and found: “For a long time, geneticists studying this large group puzzled over its evolutionary history, with different studies finding different relationships that seemed to conflict with each other.”
The article closes by explaining that nice, neat evolutionary trees are not accurate: “I imagine having the chance to speak to my students again. I would place Haeckel’s tree on the screen and tell them that it is an anachronism.”
Testing the Test
I can’t conclude before remarking on the validity of Dawkins’s test of intelligent design. In some ways it is dubious, but in other ways it makes some sense. He’s probably right that “perfect” congruency among trees is not best explained by intelligent design. But ID as a scientific theory does not claim that everything must be the result of design. After all, the most that perfect phylogenetic congruency would demonstrate is common ancestry — an idea that’s accepted by some proponents of intelligent design because the original design could have been located in the common ancestor!
Also, Dawkins would be wrong to claim that any degree of treelike distribution of genes refutes intelligent design. This is because designed structures are loaded with functions, and functional components often depend upon and interact with other functional components in logical, functionally necessary ways. This can lead to non-random correlations and hierarchical distributions of traits — i.e., design can give you some treelike structure in a dataset!
For example, when you find buttons on a shirt, you’re also going to find buttonholes. When you find a wheel on a vehicle, you’re almost always going to find an axle, too. These correlations can lead to a treelike distribution of traits. Physicist Brian Miller explains this concept in a recent volume, Science and Faith in Dialogue:
Design architectures often fall into a hierarchical pattern. All transportation vehicles have certain common features such as allocated space for cargo and or passengers, propulsion system and steering. Cars possess all these features plus such components as wheels, breaks, coolants, lubricants and axles. Toyota Camry models possess all these features plus additional specialised components. The similarities in transportation vehicles would likely fit into a constructed tree at least as well as different groups of species.
There are undoubtedly many analogous correlations between biological traits —which lead to some treelike structure to a dataset. For example, genes for noses in mammals will probably also be found with genes for nose hairs. And genes for toes in terrestrial tetrapods often correlate with genes for toenails! But intelligent agents are not compelled to distribute traits as a rule according to a nested hierarchy. Thus, as Dr. Miller explains, finding some treelike structure but also lots of non-treelike distribution of traits is a reasonable expectation of design:
While many features in human products fit into a hierarchical tree-like pattern, many break that pattern. A police car and an airplane both have two-way radios while two-way radios are absent in most other cars. In addition, the same circuitry is implemented in a wide variety of vehicles to meet similar goals. This pattern reflects how engineers often create modules that can be used in diverse contexts. The modules must be designed with the explicit intent of operating in different products, and the products that use the modules must be designed to properly incorporate them into their operations. The same pattern and constraints are observed in life.
Computer scientist Winston Ewert is even developing a model of understanding “common design” through dependency graphs.
A further note: In the 2009 interview, Dawkins’s refers to “genes that are no longer functional,” aka so-called pseudogenes. He argues that this refutes intelligent design because no designer would put identical non-functional genes in the same locations of the genomes of different species. But this is a weak argument given that so many pseudogenes are now known to have function. Shared pseudogenes that truly are nonfunctional are probably better explained by common ancestry than by common design. That said, even if they are nonfunctional today, it’s possible that such pseudogenes were originally designed to be functional but their functions degraded over time so that they are “no longer functional.”
This is again where design and common ancestry could intersect: shared pseudogenes — if truly nonfunctional — may have been originally designed as functional, but they would most likely have been designed in the common ancestor rather than designed in species separately. Dawkins’s point about pseudogenes is not a test of design on the grand scale but only a test of separate yet common design as compared with common ancestry (which could still potentially involve design).
What Will Richard Dawkins Say Now?
In any case, it’s true that the papers and articles I cited above were not written to support intelligent design. However, they affirm that there are frequent conflicts between phylogenetic trees, and they affirm non-treelike data among different genes and different species. This conclusion doesn’t come only from old papers or papers studying the tips of the tree of life, but from recent studies looking at key aspects of that tree, such as fundamental animal or plant or microorganismal relationships. And this frequent incongruity and discordance between phylogenetic trees is exactly what Dawkins says is predicted by intelligent design.
Dawkins has set up a test for Darwinian evolution versus intelligent design. He needs to live with his own repeated statements. Will he admit that his prediction has failed for evolution but succeeded for intelligent design?
